You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001385_00221
Basic Information
help
| Species |
Alistipes_A indistinctus
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A indistinctus
|
| CAZyme ID |
MGYG000001385_00221
|
| CAZy Family |
GH106 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001385 |
2850981 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 262234;
End: 265122
Strand: -
|
No EC number prediction in MGYG000001385_00221.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
46 |
960 |
5.6e-192 |
0.9951456310679612 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam17132
|
Glyco_hydro_106 |
1.48e-40 |
148 |
770 |
284 |
866 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 6Q2F_A
|
1.70e-31 |
210 |
962 |
410 |
1141 |
Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A
|
6.93e-29 |
208 |
959 |
362 |
1098 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A
|
1.58e-28 |
208 |
959 |
362 |
1098 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
This protein is predicted as SP
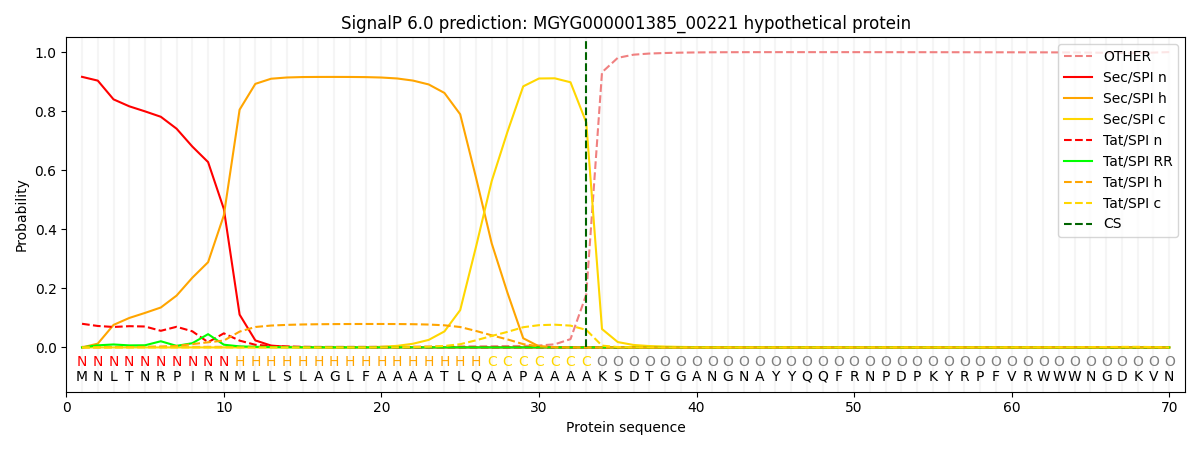
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.004330
|
0.910387
|
0.002107
|
0.082259
|
0.000582
|
0.000304
|
There is no transmembrane helices in MGYG000001385_00221.

