You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001385_01154
You are here: Home > Sequence: MGYG000001385_01154
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes_A indistinctus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes_A; Alistipes_A indistinctus | |||||||||||
| CAZyme ID | MGYG000001385_01154 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1435879; End: 1438956 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 268 | 1021 | 6.7e-208 | 0.883495145631068 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 62 | 820 | 2 | 870 | alpha-L-rhamnosidase. |
| pfam02837 | Glyco_hydro_2_N | 6.76e-04 | 866 | 987 | 33 | 144 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCG54391.1 | 0.0 | 43 | 1025 | 1 | 983 |
| AWO01984.1 | 0.0 | 50 | 1025 | 14 | 990 |
| BAU55722.1 | 7.11e-315 | 45 | 1019 | 18 | 1000 |
| SCM59048.1 | 3.92e-310 | 50 | 1025 | 21 | 1001 |
| ASU36259.1 | 3.34e-307 | 55 | 1025 | 17 | 994 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.71e-129 | 15 | 1021 | 4 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.43e-101 | 60 | 1019 | 35 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 3.74e-101 | 60 | 1019 | 35 | 1096 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 8.16e-156 | 51 | 1021 | 25 | 1155 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
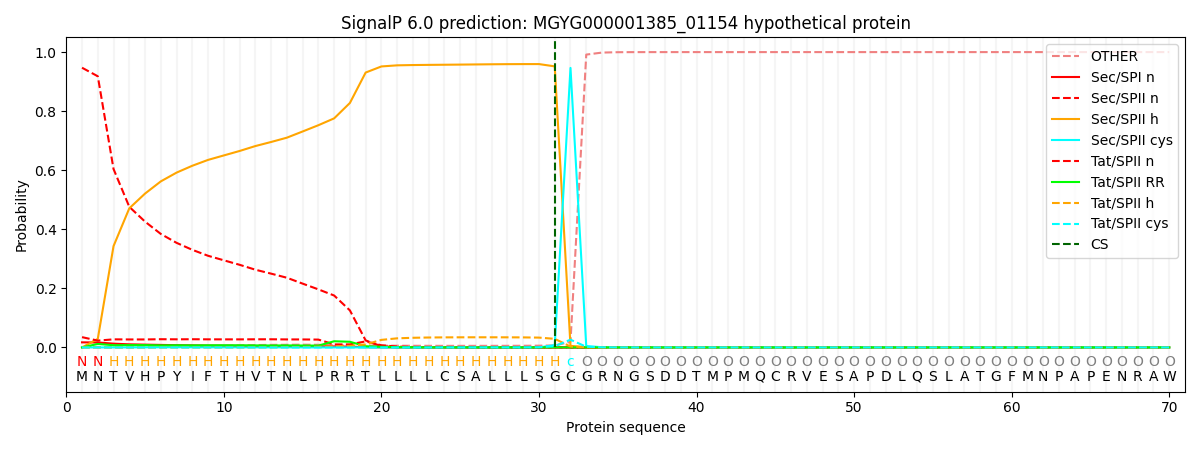
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000228 | 0.004656 | 0.960298 | 0.000063 | 0.034764 | 0.000002 |
