You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001386_03514
Basic Information
help
| Species |
Enterocloster clostridioformis
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster clostridioformis
|
| CAZyme ID |
MGYG000001386_03514
|
| CAZy Family |
GH26 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 345 |
|
40070.4 |
4.0368 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001386 |
5455983 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 91076;
End: 92113
Strand: -
|
No EC number prediction in MGYG000001386_03514.
| Family |
Start |
End |
Evalue |
family coverage |
| GH26 |
150 |
294 |
2.2e-24 |
0.42574257425742573 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG4124
|
ManB2 |
4.54e-16 |
164 |
299 |
177 |
318 |
Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156
|
Glyco_hydro_26 |
3.51e-09 |
170 |
281 |
156 |
258 |
Glycosyl hydrolase family 26. |
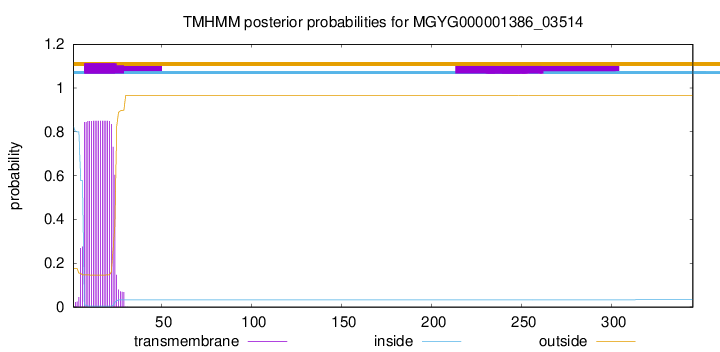
This protein is predicted as LIPO
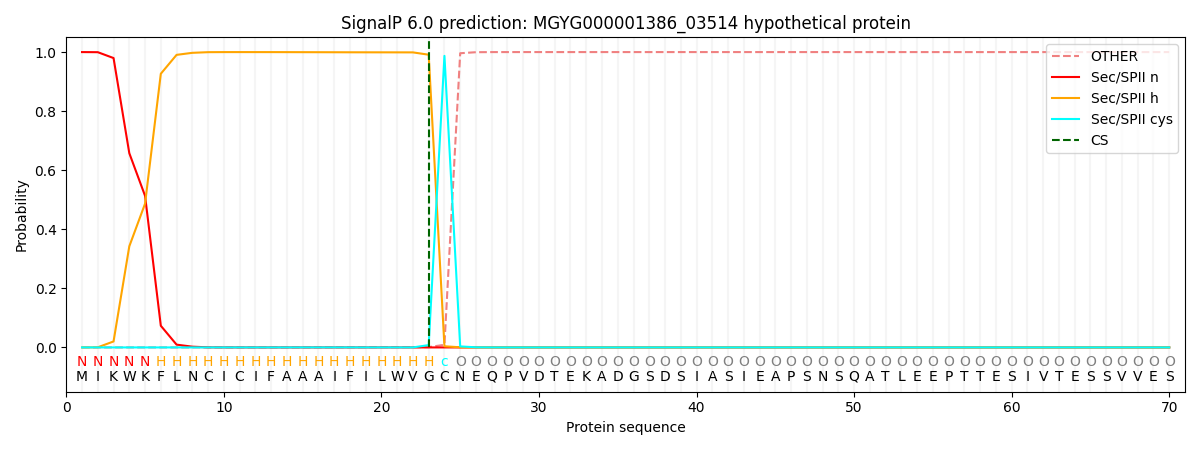
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000000
|
0.000002
|
1.000053
|
0.000000
|
0.000000
|
0.000000
|