You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001391_00464
You are here: Home > Sequence: MGYG000001391_00464
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprobacter fastidiosus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter fastidiosus | |||||||||||
| CAZyme ID | MGYG000001391_00464 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 526579; End: 529848 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 888 | 1003 | 2.2e-19 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.52e-12 | 887 | 1002 | 4 | 126 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam12708 | Pectate_lyase_3 | 4.48e-12 | 42 | 108 | 3 | 65 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam12708 | Pectate_lyase_3 | 4.57e-11 | 473 | 527 | 6 | 61 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG5434 | Pgu1 | 8.97e-07 | 474 | 524 | 88 | 137 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| COG5434 | Pgu1 | 1.39e-06 | 32 | 143 | 74 | 178 | Polygalacturonase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHW60575.1 | 0.0 | 26 | 1009 | 1 | 972 |
| AEV98037.1 | 0.0 | 7 | 1005 | 13 | 1004 |
| QUT90069.1 | 1.06e-306 | 23 | 1089 | 27 | 1120 |
| QJD86468.1 | 4.70e-86 | 23 | 785 | 906 | 1666 |
| QHW29527.1 | 1.16e-68 | 27 | 843 | 1086 | 1992 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XLR_A | 2.51e-07 | 875 | 999 | 5 | 139 | ChainA, Galactose oxidase [Fusarium graminearum],6XLS_A Chain A, Galactose oxidase [Fusarium graminearum] |
| 6XLT_A | 2.51e-07 | 875 | 999 | 5 | 139 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 2WQ8_A | 2.54e-07 | 875 | 999 | 26 | 160 | ChainA, GALACTOSE OXIDASE [Fusarium graminearum] |
| 5LW3_A | 2.86e-07 | 473 | 642 | 7 | 187 | Azotobactervinelandii Mannuronan C-5 epimerase AlgE6 A-module [Azotobacter vinelandii] |
| 1GOF_A | 3.28e-07 | 875 | 999 | 4 | 138 | NOVELTHIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49426 | 3.85e-07 | 42 | 539 | 72 | 491 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
| I1S2N3 | 1.41e-06 | 875 | 999 | 45 | 179 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0CS93 | 1.85e-06 | 875 | 999 | 45 | 179 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
| Q9ZFH0 | 2.69e-06 | 473 | 642 | 7 | 187 | Mannuronan C5-epimerase AlgE6 OS=Azotobacter vinelandii OX=354 GN=algE6 PE=1 SV=1 |
| Q9ZFG9 | 6.05e-06 | 473 | 527 | 7 | 68 | Alginate lyase 7 OS=Azotobacter vinelandii OX=354 GN=algE7 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
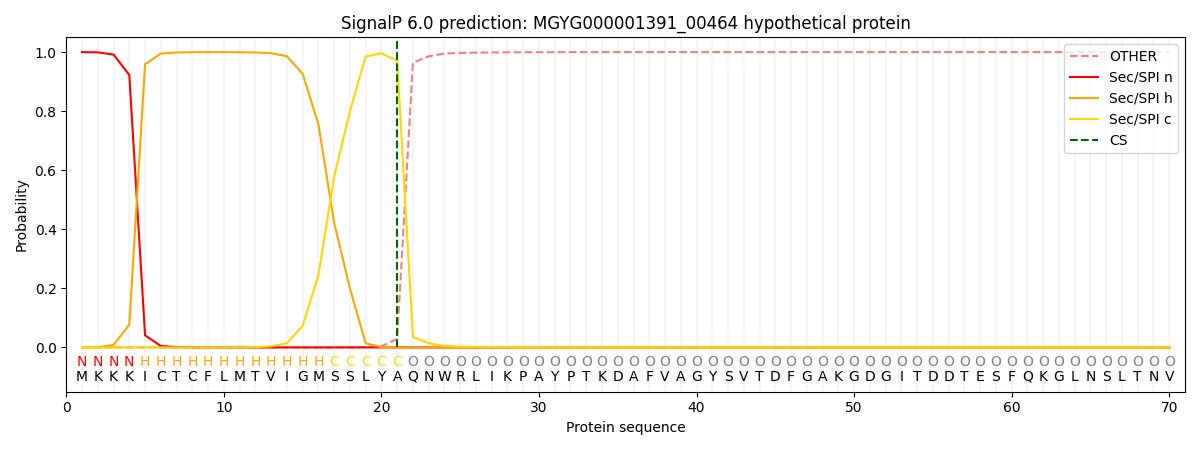
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000939 | 0.998189 | 0.000270 | 0.000209 | 0.000193 | 0.000188 |
