You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001391_02456
You are here: Home > Sequence: MGYG000001391_02456
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
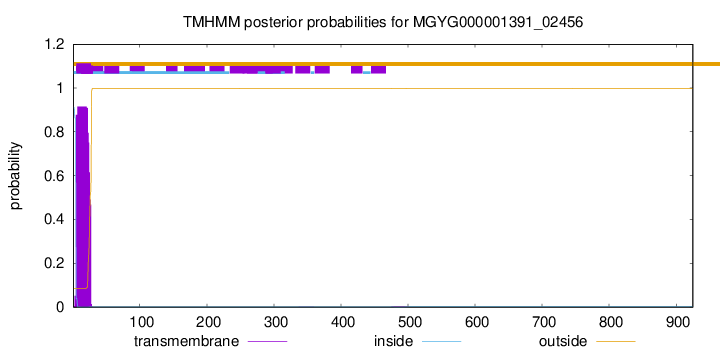
TMHMM annotations
Basic Information help
| Species | Coprobacter fastidiosus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter fastidiosus | |||||||||||
| CAZyme ID | MGYG000001391_02456 | |||||||||||
| CAZy Family | CBM6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14169; End: 16946 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM6 | 546 | 682 | 2e-25 | 0.9855072463768116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16216 | GxGYxYP_N | 1.60e-60 | 58 | 281 | 5 | 215 | GxGYxY sequence motif in domain of unknown function N-terminal. This domain is found in bacteria, archaea and eukaryotes, and is typically between 213 and 231 amino acids in length. This domain is found in association with pfam14323. |
| cd04080 | CBM6_cellulase-like | 5.08e-41 | 540 | 682 | 1 | 144 | Carbohydrate Binding Module 6 (CBM6); appended to glycoside hydrolase (GH) domains, including GH5 (cellulase). This family includes carbohydrate binding module 6 (CBM6) domains that are appended to several glycoside hydrolase (GH) domains, including GH5 (cellulase) and GH16, as well as to coagulation factor 5/8 carbohydrate-binding domains. CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. The CBM6s are appended to GHs that display a diversity of substrate specificities. For some members of this family information is available about the specific substrates of the appended GH domains. It includes the CBM domains of various enzymes involved in cell wall degradation including, an extracellular beta-1,3-glucanase from Lysobacter enzymogenes encoded by the gluC gene (its catalytic domain belongs to the GH16 family), the tandem CBM domains of Pseudomonas sp. PE2 beta-1,3(4)-glucanase A (its catalytic domain also belongs to GH16), and a family 6 CBM from Cellvibrio mixtus Endoglucanase 5A (CmCBM6) which binds to the beta1,4-beta1,3-mixed linked glucans lichenan, and barley beta-glucan, cello-oligosaccharides, insoluble forms of cellulose, the beta1,3-glucan laminarin, and xylooligosaccharides, and the CBM6 of Fibrobacter succinogenes S85 XynD xylanase, appended to a GH10 domain, and Cellvibrio japonicas Cel5G appended to a GH5 (cellulase) domain. GH5 (cellulase) family includes enzymes with several known activities such as endoglucanase, beta-mannanase, and xylanase, which are involved in the degradation of cellulose and xylans. GH16 family includes enzymes with lichenase, xyloglucan endotransglycosylase (XET), and beta-agarase activities. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. For CmCBM6 it has been shown that these two binding sites have different ligand specificities. |
| pfam14323 | GxGYxYP_C | 2.76e-30 | 304 | 484 | 1 | 200 | GxGYxYP putative glycoside hydrolase C-terminal domain. This family carries a characteristic sequence motif, GxGYxYP, and is a putative glycoside hydrolase. This domain is found in association with pfam16216. Associated families are sugar-processing domains. |
| smart00606 | CBD_IV | 4.44e-14 | 539 | 681 | 3 | 129 | Cellulose Binding Domain Type IV. |
| cd04079 | CBM6_agarase-like | 1.68e-11 | 543 | 668 | 5 | 116 | Carbohydrate Binding Module 6 (CBM6); appended mainly to glycoside hydrolase (GH) family 16 alpha- and beta agarases. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family 16 agarases. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the activity of alpha- and beta-agarase catalytic modules which are involved in the hydrolysis of 1,4-beta-D-galactosidic linkages. These CBM6s bind specifically to the non-reducing end of agarose chains, recognizing only the first repeat of the disaccharide, and directing the appended catalytic modules to areas of the plant cell wall attacked by beta-agarases. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. This family includes three tandem CBM6s from the Saccharophagus degradans agarase Aga86E, and three tandem CBM6s from Vibrio sp. strain PO-303 AgaA; in both these proteins these are appended to a GH16 domain. Vibrio AgaA also contains a Big-2-like protein-protein interaction domain. This family also includes two tandem CBM6s from an endo-type beta-agarase from a deep-sea Microbulbifer-like isolate, which are appended to a GH16 domain, and two of three CBM6s of Alteromonas agarilytica AgaA alpha-agarase, which are appended to a GH96 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT77716.1 | 0.0 | 27 | 925 | 27 | 914 |
| ANH82740.1 | 5.74e-29 | 56 | 377 | 74 | 380 |
| BCJ99038.1 | 8.49e-27 | 38 | 404 | 180 | 536 |
| ACL15778.1 | 2.37e-26 | 450 | 689 | 160 | 390 |
| AXT61911.1 | 2.01e-25 | 529 | 740 | 236 | 441 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5G56_A | 5.10e-24 | 543 | 808 | 340 | 598 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 3SGG_A | 5.78e-18 | 67 | 404 | 79 | 400 | Crystalstructure of a putative hydrolase (BT_2193) from Bacteroides thetaiotaomicron VPI-5482 at 1.25 A resolution [Bacteroides thetaiotaomicron] |
| 2Y8K_A | 6.18e-18 | 543 | 680 | 342 | 479 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5LA0_A Chain A, Carbohydrate binding family 6 [Acetivibrio thermocellus JW20],5LA1_A Chain A, Carbohydrate binding family 6 [Acetivibrio thermocellus JW20] |
| 5LA2_A | 6.18e-18 | 543 | 680 | 342 | 479 | ChainA, Carbohydrate binding family 6 [Acetivibrio thermocellus],5LA2_B Chain B, Carbohydrate binding family 6 [Acetivibrio thermocellus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
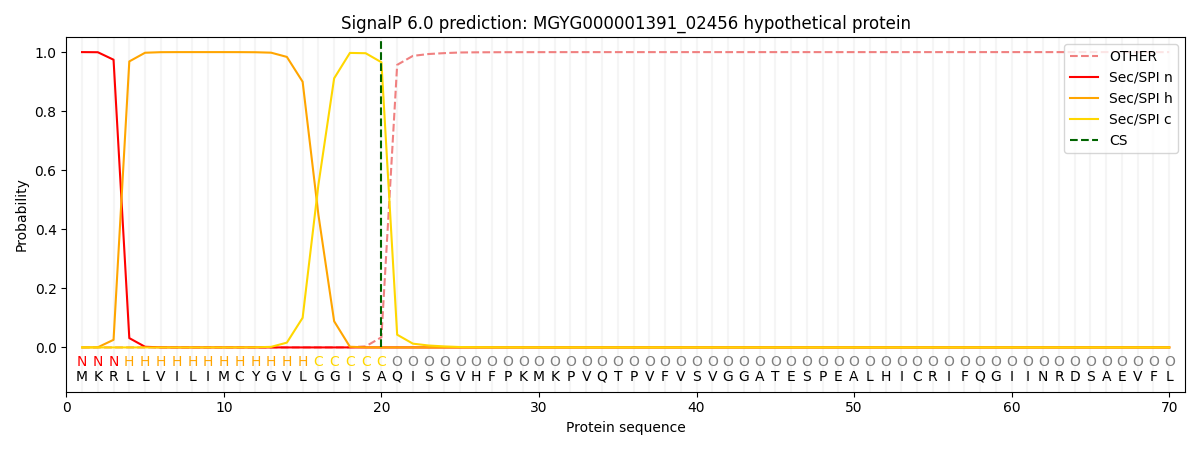
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000414 | 0.998898 | 0.000185 | 0.000165 | 0.000157 | 0.000150 |