You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001394_03864
You are here: Home > Sequence: MGYG000001394_03864
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Yokenella regensburgei | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Yokenella; Yokenella regensburgei | |||||||||||
| CAZyme ID | MGYG000001394_03864 | |||||||||||
| CAZy Family | AA10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1938533; End: 1939342 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA10 | 39 | 267 | 4.4e-35 | 0.9943820224719101 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21178 | Fusolin-like | 4.95e-60 | 39 | 266 | 1 | 225 | fusolin and similar proteins. Fusolin is a protein found in spindles of insect poxviruses that resembles the lytic polysaccharide monooxygenases of chitinovorous bacteria and may function to disrupt the chitin-rich peritrophic matrix that protects insects against oral infections. Thus, it is a component of the virus occlusion bodies (which are large proteinaceous polyhedra) that protect the virus from the outside environment for extended periods until they are ingested by insect larvae. |
| PHA03387 | gp37 | 1.44e-51 | 39 | 266 | 20 | 248 | spherodin-like protein; Provisional |
| cd21177 | LPMO_AA10 | 1.45e-48 | 39 | 267 | 1 | 180 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| pfam03067 | LPMO_10 | 4.05e-38 | 39 | 266 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| COG3397 | COG3397 | 6.40e-16 | 34 | 268 | 25 | 207 | Predicted carbohydrate-binding protein, contains CBM5 and CBM33 domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHA89360.1 | 1.62e-131 | 18 | 269 | 17 | 261 |
| AML58481.1 | 7.04e-124 | 1 | 269 | 1 | 263 |
| VEI70086.1 | 4.04e-85 | 107 | 269 | 1 | 161 |
| QBX64745.1 | 4.90e-81 | 30 | 269 | 27 | 275 |
| QGH63042.1 | 4.90e-81 | 8 | 269 | 8 | 275 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN2_A | 5.65e-32 | 39 | 267 | 1 | 229 | THEATOMIC STRUCTURE OF WISEANA SPP ENTOMOPOXVIRUS (WSEPV) FUSOLIN SPINDLES [unidentified entomopoxvirus] |
| 4YN1_A | 1.31e-29 | 39 | 265 | 1 | 230 | THEATOMIC STRUCTURE OF ANOMALA CUPREA ENTOMOPOXVIRUS (ACEPV) FUSOLIN SPINDLES [Anomala cuprea entomopoxvirus] |
| 4X27_A | 4.21e-28 | 39 | 266 | 1 | 231 | Structuralbasis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles (complex with Copper) [Entomopoxvirinae],4X29_A Structural basis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles (complex with Zinc) [Entomopoxvirinae] |
| 4OW5_A | 4.28e-28 | 39 | 266 | 1 | 231 | Structuralbasis for the enhancement of virulence by entomopoxvirus fusolin and its in vivo crystallization into viral spindles [unidentified entomopoxvirus] |
| 6NDQ_A | 1.42e-08 | 129 | 267 | 62 | 184 | Crystalstructure of a LPMO from Kitasatospora papulosa [[Kitasatospora] papulosa],6NDQ_B Crystal structure of a LPMO from Kitasatospora papulosa [[Kitasatospora] papulosa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q05894 | 1.51e-34 | 39 | 266 | 21 | 251 | Spindolin OS=Heliothis armigera entomopoxvirus OX=10290 PE=1 SV=1 |
| P23061 | 2.53e-33 | 29 | 266 | 11 | 252 | Spindolin OS=Choristoneura biennis entomopoxvirus OX=10288 GN=SPH PE=1 SV=1 |
| Q65328 | 1.42e-29 | 39 | 266 | 19 | 246 | Spheroidin-like protein OS=Orgyia pseudotsugata multicapsid polyhedrosis virus OX=262177 GN=SLP PE=2 SV=1 |
| P23058 | 1.93e-29 | 39 | 266 | 20 | 247 | Spheroidin-like protein OS=Autographa californica nuclear polyhedrosis virus OX=46015 GN=SLP PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
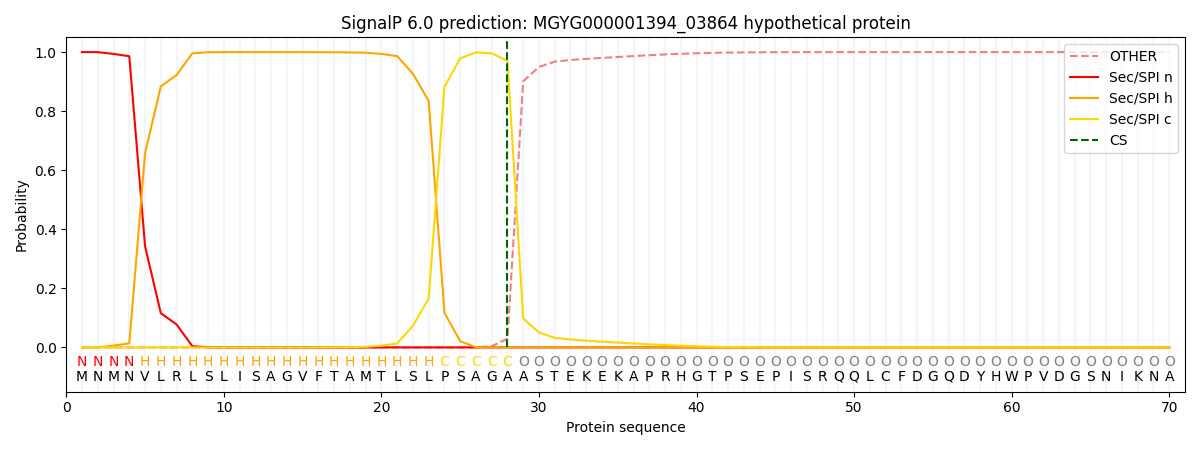
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000383 | 0.998857 | 0.000179 | 0.000224 | 0.000174 | 0.000154 |
