You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001395_01038
You are here: Home > Sequence: MGYG000001395_01038
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
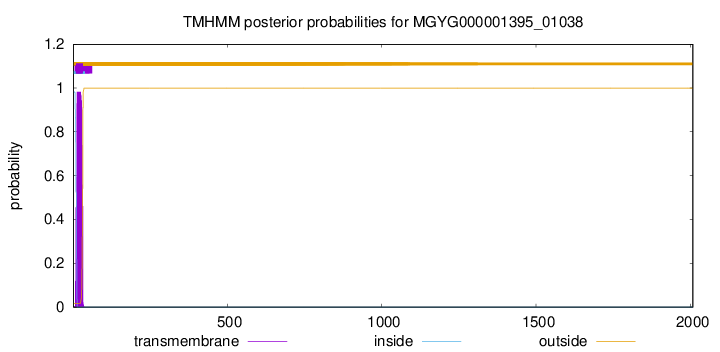
TMHMM annotations
Basic Information help
| Species | Listeria innocua | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria innocua | |||||||||||
| CAZyme ID | MGYG000001395_01038 | |||||||||||
| CAZy Family | CBM16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 202136; End: 208162 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3209 | RhsA | 5.79e-48 | 954 | 2001 | 4 | 773 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| NF033679 | DNRLRE_dom | 8.46e-27 | 295 | 458 | 1 | 164 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
| COG3209 | RhsA | 1.39e-09 | 861 | 1108 | 38 | 316 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| pfam05593 | RHS_repeat | 1.22e-05 | 1794 | 1824 | 1 | 31 | RHS Repeat. RHS proteins contain extended repeat regions. These repeats often appear to be involved in ligand binding. Note that this model may not find all the repeats in a protein and that it covers two RHS repeats. The 3D structure of an RHS-repeat-containing protein (the B and C components of an ABC toxin complex) has been determined. The RHS repeats form an extended strip of beta-sheet that spirals around to form a hollow shell, encapsulating the variable C-terminal domain. |
| COG3209 | RhsA | 6.40e-05 | 867 | 1129 | 225 | 534 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGF42541.1 | 0.0 | 1 | 2001 | 1 | 2001 |
| AUI60804.1 | 7.49e-25 | 50 | 479 | 45 | 463 |
| ANN19275.1 | 8.69e-24 | 18 | 479 | 1 | 450 |
| QRT48460.1 | 6.62e-22 | 1516 | 1985 | 1779 | 2243 |
| AEV81245.1 | 5.25e-20 | 64 | 525 | 163 | 618 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q07833 | 8.19e-312 | 32 | 2001 | 55 | 2126 | tRNA nuclease WapA OS=Bacillus subtilis (strain 168) OX=224308 GN=wapA PE=1 SV=2 |
| G4NYJ6 | 3.45e-309 | 25 | 2001 | 48 | 2126 | tRNA3(Ser)-specific nuclease WapA OS=Bacillus spizizenii (strain DSM 15029 / JCM 12233 / NBRC 101239 / NRRL B-23049 / TU-B-10) OX=1052585 GN=wapA PE=1 SV=1 |
| D4G3R4 | 1.66e-299 | 32 | 2001 | 68 | 2139 | tRNA(Glu)-specific nuclease WapA OS=Bacillus subtilis subsp. natto (strain BEST195) OX=645657 GN=wapA PE=1 SV=2 |
| P42018 | 8.26e-28 | 40 | 244 | 36 | 245 | Wall-associated protein OS=Geobacillus stearothermophilus OX=1422 GN=wapA' PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
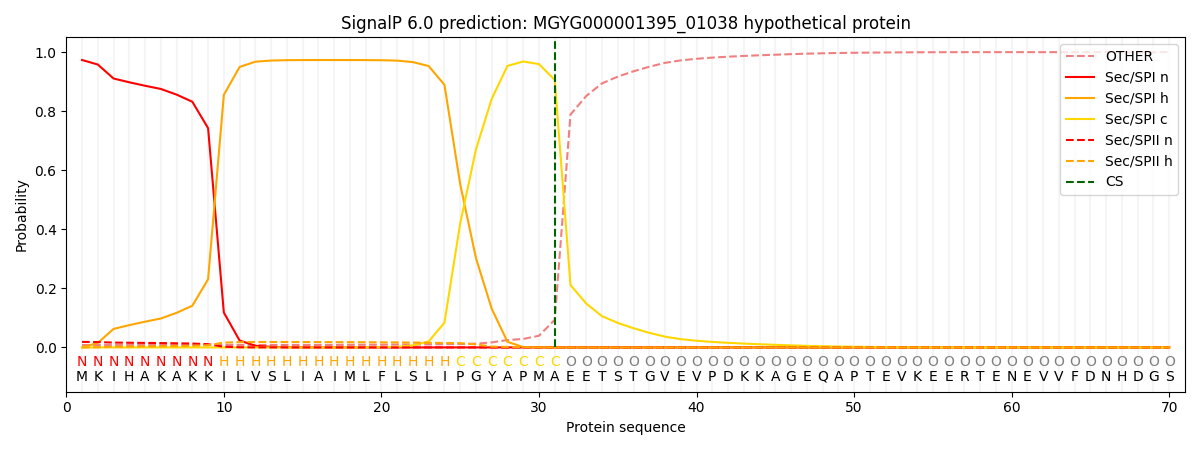
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.010203 | 0.968473 | 0.020366 | 0.000414 | 0.000260 | 0.000236 |