You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001395_01221
You are here: Home > Sequence: MGYG000001395_01221
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
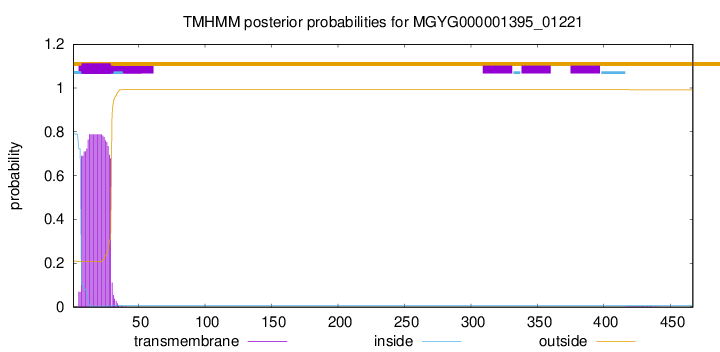
TMHMM annotations
Basic Information help
| Species | Listeria innocua | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria innocua | |||||||||||
| CAZyme ID | MGYG000001395_01221 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | putative endopeptidase p60 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 380795; End: 382198 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13914 | PRK13914 | 0.0 | 1 | 467 | 3 | 481 | invasion associated endopeptidase. |
| pfam00877 | NLPC_P60 | 4.21e-48 | 364 | 466 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG3103 | YgiM | 6.77e-32 | 72 | 200 | 19 | 153 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| COG0791 | Spr | 4.65e-31 | 350 | 464 | 73 | 196 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| NF033742 | NlpC_p60_RipB | 1.32e-24 | 356 | 448 | 81 | 185 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CAC95823.1 | 1.23e-238 | 1 | 467 | 1 | 465 |
| QUR86445.1 | 2.86e-238 | 1 | 467 | 1 | 469 |
| QGF42398.1 | 1.16e-237 | 1 | 467 | 1 | 469 |
| AEO72013.1 | 1.53e-237 | 1 | 467 | 3 | 467 |
| AEO72014.1 | 1.47e-236 | 1 | 467 | 3 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HPE_A | 5.08e-22 | 358 | 467 | 193 | 306 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 7CFL_A | 4.99e-21 | 354 | 448 | 16 | 115 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 6B8C_A | 1.75e-17 | 354 | 446 | 30 | 123 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 3I86_A | 2.78e-15 | 355 | 448 | 15 | 120 | Crystalstructure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 [Mycobacterium avium subsp. paratuberculosis],3I86_B Crystal structure of the P60 Domain from M. avium subspecies paratuberculosis antigen MAP1204 [Mycobacterium avium subsp. paratuberculosis] |
| 3PBC_A | 2.90e-15 | 355 | 448 | 88 | 193 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01836 | 2.64e-239 | 1 | 467 | 3 | 467 | Probable endopeptidase p60 OS=Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) OX=272626 GN=iap PE=3 SV=2 |
| P21171 | 4.14e-185 | 1 | 467 | 3 | 484 | Probable endopeptidase p60 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=iap PE=1 SV=2 |
| Q01839 | 1.63e-169 | 1 | 467 | 3 | 524 | Probable endopeptidase p60 OS=Listeria welshimeri OX=1643 GN=iap PE=3 SV=1 |
| Q01838 | 6.29e-164 | 1 | 467 | 3 | 523 | Probable endopeptidase p60 OS=Listeria seeligeri OX=1640 GN=iap PE=3 SV=1 |
| Q01837 | 1.30e-163 | 1 | 467 | 3 | 524 | Probable endopeptidase p60 OS=Listeria ivanovii OX=1638 GN=iap PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
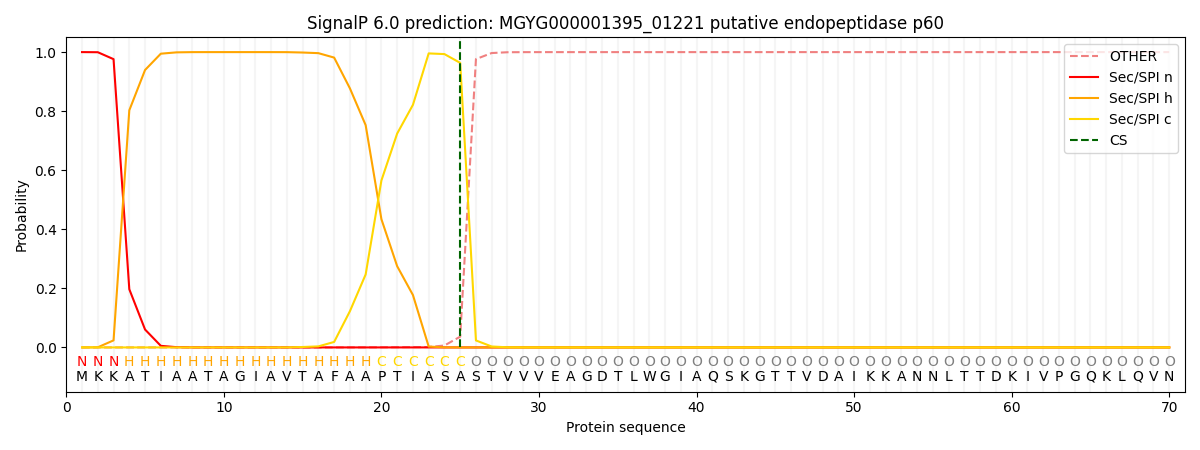
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000233 | 0.999041 | 0.000168 | 0.000194 | 0.000177 | 0.000154 |