You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001399_02790
You are here: Home > Sequence: MGYG000001399_02790
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Odoribacter laneus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Odoribacter; Odoribacter laneus | |||||||||||
| CAZyme ID | MGYG000001399_02790 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 365347; End: 366987 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 9 | 426 | 1.2e-60 | 0.7833333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 3.70e-31 | 9 | 404 | 11 | 412 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 3.69e-14 | 29 | 394 | 31 | 397 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 3.76e-12 | 62 | 223 | 2 | 160 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| cd07343 | M48A_Zmpste24p_like | 0.002 | 293 | 433 | 63 | 197 | Peptidase M48 subfamily A, a type 1 CaaX endopeptidase. This family contains peptidase family M48 subfamily A which includes a number of well-characterized genes such as those found in humans (ZMPSTE24, also known as farnesylated protein-converting enzyme 1 or FACE-1 or Hs Ste24), Taenia solium metacestode (TsSte24p), Arabidopsis (AtSte24) and yeast (Ste24p). Ste24p contains the zinc metalloprotease motif (HEXXH), likely exposed on the cytoplasmic side. It is thought to be intimately associated with the endoplasmic reticulum (ER), regardless of whether its genes possess the conventional signal motif (KKXX) in the C-terminal. Proteins in this family proteolytically remove the C-terminal three residues of farnesylated proteins. Ste24p is involved in the post-translational processing of prelamin A to mature lamin A, a major component of the nuclear envelope. ZmpSte24 deficiency causes an accumulation of prelamin A leading to lipodystrophy and other disease phenotypes, while mutations in this gene or in that encoding its substrate, prelamin A, result in a series of human inherited diseases known as laminopathies, the most severe of which are Hutchinson Gilford progeria syndrome (HGPS) and restrictive dermopathy (RD) which arise due to unsuccessful maturation of prelamin A. Two forms of mandibuloacral dysplasia, a condition that causes a variety of abnormalities involving bone development, skin pigmentation, and fat distribution, are caused by mutations in two different genes; mutations in the LMNA gene, which normally provides instructions for making lamin A and lamin C, cause mandibuloacral dysplasia with A-type lipodystrophy (MAD-A), and mutations in the ZMPSTE24 gene cause mandibuloacral dysplasia with B-type lipodystrophy (MAD-B). Within cells, these genes are involved in maintaining the structure of the nucleus and may play a role in many cellular processes. Certain HIV protease inhibitors have been shown to inhibit the enzymatic activity of ZMPSTE24, but not enzymes involved in prelamin A processing. |
| COG4346 | COG4346 | 0.006 | 61 | 150 | 135 | 223 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZO94843.1 | 3.41e-19 | 29 | 330 | 38 | 338 |
| QTL99938.1 | 4.53e-19 | 29 | 330 | 38 | 338 |
| BAT71475.1 | 6.67e-19 | 25 | 366 | 25 | 350 |
| QTL97755.1 | 7.78e-19 | 10 | 414 | 27 | 438 |
| ADR19199.1 | 1.00e-18 | 4 | 318 | 6 | 316 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O67270 | 8.76e-14 | 23 | 317 | 22 | 308 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| Q3KCC9 | 1.04e-13 | 13 | 426 | 27 | 438 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1 |
| A1JPM4 | 1.33e-13 | 29 | 348 | 32 | 352 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Yersinia enterocolitica serotype O:8 / biotype 1B (strain NCTC 13174 / 8081) OX=393305 GN=arnT PE=3 SV=1 |
| A4SQX1 | 2.89e-12 | 24 | 363 | 23 | 361 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas salmonicida (strain A449) OX=382245 GN=arnT PE=3 SV=1 |
| A0KGY4 | 3.82e-12 | 17 | 363 | 11 | 361 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Aeromonas hydrophila subsp. hydrophila (strain ATCC 7966 / DSM 30187 / BCRC 13018 / CCUG 14551 / JCM 1027 / KCTC 2358 / NCIMB 9240 / NCTC 8049) OX=380703 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000033 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
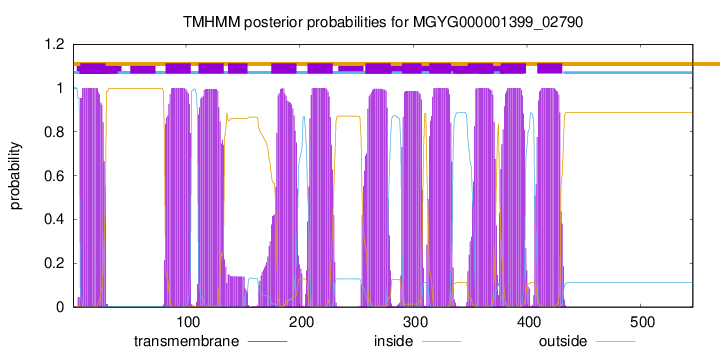
| start | end |
|---|---|
| 7 | 29 |
| 82 | 104 |
| 111 | 133 |
| 137 | 154 |
| 175 | 197 |
| 207 | 229 |
| 258 | 280 |
| 290 | 307 |
| 314 | 333 |
| 348 | 370 |
| 377 | 399 |
| 409 | 431 |
