You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001402_01375
You are here: Home > Sequence: MGYG000001402_01375
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprobacillus cateniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Coprobacillus; Coprobacillus cateniformis | |||||||||||
| CAZyme ID | MGYG000001402_01375 | |||||||||||
| CAZy Family | CBM40 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1355965; End: 1359780 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01834 | SGNH_hydrolase_like_2 | 8.98e-24 | 677 | 876 | 4 | 191 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 7.41e-22 | 679 | 866 | 1 | 174 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 9.56e-20 | 677 | 875 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| cd04501 | SGNH_hydrolase_like_4 | 8.26e-16 | 678 | 877 | 4 | 183 | Members of the SGNH-hydrolase superfamily, a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| cd01822 | Lysophospholipase_L1_like | 6.50e-12 | 678 | 869 | 4 | 168 | Lysophospholipase L1-like subgroup of SGNH-hydrolases. The best characterized member in this family is TesA, an E. coli periplasmic protein with thioesterase, esterase, arylesterase, protease and lysophospholipase activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANU71609.1 | 5.75e-124 | 342 | 984 | 1672 | 2325 |
| ASU26373.1 | 5.75e-124 | 342 | 984 | 1672 | 2325 |
| QQR10962.1 | 5.75e-124 | 342 | 984 | 1672 | 2325 |
| ARV02485.1 | 5.75e-124 | 342 | 984 | 1672 | 2325 |
| QQD64786.1 | 1.70e-123 | 304 | 963 | 1485 | 2142 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3RJT_A | 1.02e-06 | 734 | 880 | 63 | 216 | Crystalstructure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 [Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446],3RJT_B Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 [Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P75377 | 1.55e-13 | 356 | 779 | 107 | 640 | Uncharacterized protein MPN_407 OS=Mycoplasma pneumoniae (strain ATCC 29342 / M129) OX=272634 GN=MPN_407 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
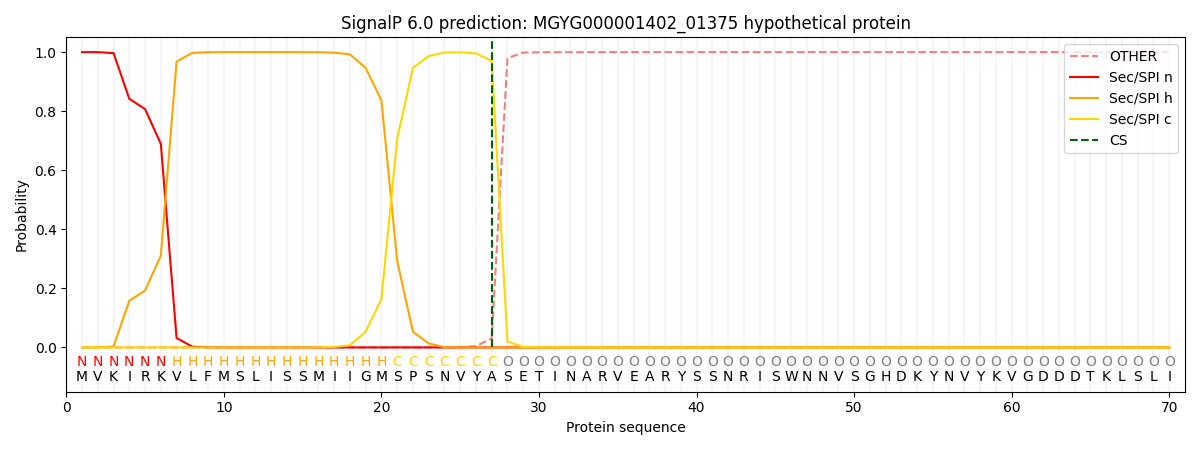
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000587 | 0.998518 | 0.000213 | 0.000249 | 0.000203 | 0.000179 |
