You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001402_02662
You are here: Home > Sequence: MGYG000001402_02662
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
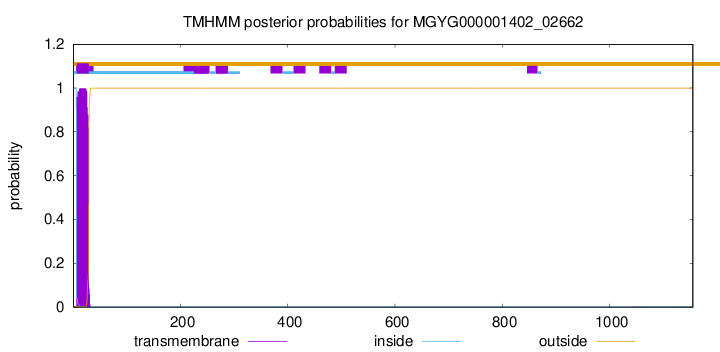
TMHMM annotations
Basic Information help
| Species | Coprobacillus cateniformis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Coprobacillus; Coprobacillus cateniformis | |||||||||||
| CAZyme ID | MGYG000001402_02662 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2698793; End: 2702260 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 799 | 1065 | 1.1e-62 | 0.4337837837837838 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00063 | FN3 | 7.59e-11 | 320 | 406 | 1 | 90 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 9.25e-09 | 320 | 400 | 1 | 82 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| pfam00041 | fn3 | 4.02e-06 | 325 | 402 | 5 | 85 | Fibronectin type III domain. |
| pfam12708 | Pectate_lyase_3 | 3.54e-05 | 554 | 716 | 38 | 190 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG3401 | FN3 | 0.006 | 431 | 541 | 179 | 284 | Fibronectin type 3 domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLY78983.1 | 0.0 | 31 | 1145 | 30 | 1123 |
| QLY78981.1 | 0.0 | 31 | 1145 | 30 | 1129 |
| BCL56920.1 | 1.23e-302 | 149 | 1139 | 155 | 1181 |
| QMW75525.1 | 7.54e-301 | 32 | 1139 | 30 | 1161 |
| QPS14140.1 | 7.54e-301 | 32 | 1139 | 30 | 1161 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4TZ1_A | 2.91e-118 | 511 | 1103 | 9 | 532 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4TZ3_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4TZ5_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4TZ5_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4PEW_A | 3.00e-118 | 511 | 1103 | 21 | 544 | Structureof sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4PEW_B Structure of sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 4TYV_A | 3.09e-118 | 511 | 1103 | 11 | 534 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4TYV_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E] |
| 4PEX_A | 4.18e-118 | 511 | 1103 | 21 | 544 | Structureof the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEX_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEY_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4PEZ_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4PF0_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4PF0_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2NFJ9 | 1.15e-117 | 511 | 1103 | 65 | 588 | Exo-beta-1,3-glucanase OS=Streptomyces sp. (strain SirexAA-E / ActE) OX=862751 GN=SACTE_4363 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
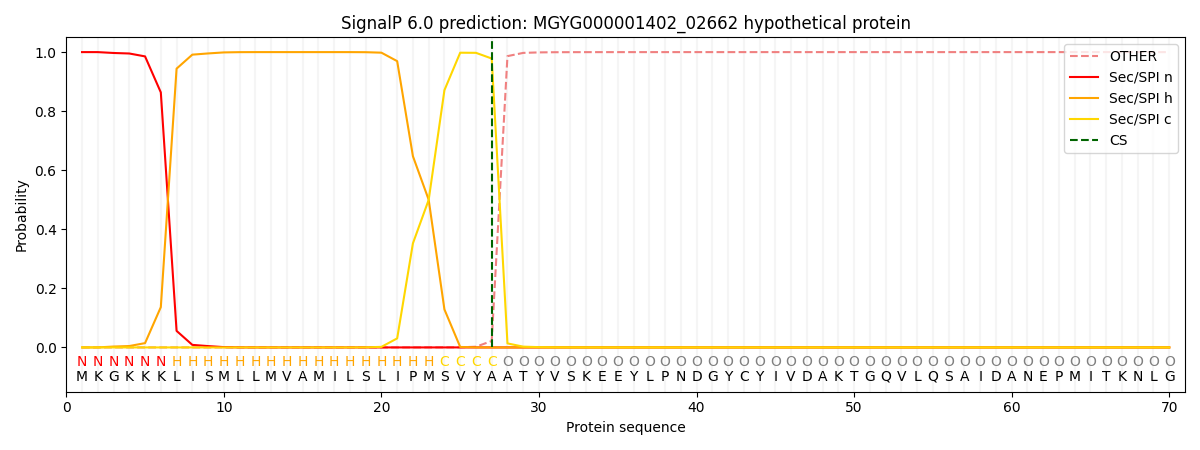
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000213 | 0.999140 | 0.000173 | 0.000166 | 0.000152 | 0.000142 |