You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001404_00194
You are here: Home > Sequence: MGYG000001404_00194
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_L senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_L; Paenibacillus_L senegalensis | |||||||||||
| CAZyme ID | MGYG000001404_00194 | |||||||||||
| CAZy Family | CBM16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 205252; End: 209136 Strand: - | |||||||||||
Full Sequence Download help
| MEWVSPIKGF RLYITIIGIV VILLGVAAFS LASAAPVNLL LNADFEKNTE HWTADSWITG | 60 |
| QEASRFTDST EETRSGSTSL MIENRVANDA KWVQKVAVEP DSYYRFSAWL KVAGANPEYT | 120 |
| GANISILGTL AMSEHYHDTG GEWTYVELVG KTGSEQTELK AALRLGGYGS LNQGKAYFED | 180 |
| VSLVKLEEAP PAAASFETEP SVATDADADS PSSKVSLVPP LLYSLAFFLL FAALYLWRIQ | 240 |
| REFLPLVRKQ SGGNDKEWFL FSLAASLLIR LLLAADMIGH PGDMATFAAW ARMAAAEGIG | 300 |
| MFYSSASFVD YPPGYIYILF VLGKLQTWFD LASGSRAFAV ILKMPSIAAD LVCAAIIYAA | 360 |
| AKRRMSGYTA LALSLLYSFN PAIIVNSAAW GQVDSFFTLF LVSGILLASR ASHGGESSSA | 420 |
| GRGRSSRANA QNRRYMLGAS AILAVALLIK PQALIFAPVY LFALYASRSW KTAASSLAVG | 480 |
| LLVFIAGIIP FTVHQEWNWI LNLYKETLQS YPYATLNAFN FFALTGANWK AQTALLFGVS | 540 |
| YQTWGWLFII GVTAFTSWLF FRRKPGVQAN YFVLALVLIA TVFVMGVKMH ERYLFPALML | 600 |
| ALMAFISTRD RRLLQLFVGF SITQFVNVCY VLLHGFEHMY AVSRLDGLLL LTSLANVLLW | 660 |
| VYLIWTAWDI FIRGNHHPIR GPKPAFSGEN AAAVEAKRAS RTGKFFSRKD WLLVSLITLI | 720 |
| YAVVALYQLG SFRAPESGWK PLASSEGVVI DLGEAYPIDR THVFSGIGSG TVTLEYSPDN | 780 |
| EQWGETQTLE LDYTRVFKWN TLSEPQDARY VRVTAVRKGF ELKEMAFFTE GSEEPIPVAL | 840 |
| VTAFGGASEE AAKALFDEQH TAVYHPTFMH STYFDEIYHA RAAYEHLDGV KPYENTHPPL | 900 |
| GKELIAAGVY LFGMNPFGWR IVGTLFGIAM IPVIYVFARR MLGKPVYGAI AAILLASDFM | 960 |
| HFAQTRIATI DVYGVFFIML MYYFMYRYYT LNFYQDGLRR TLIPLFWSGL LFGIGAASKW | 1020 |
| IVIYGGVGLA FLFFLSLYER YREYDQAKLA LASGELAGQP AIEADLRHKV SRFPGYAWAT | 1080 |
| MAWCSLFFVV IPVAVYALSF LPIMLVPGQE VSVAKLVQYQ LDMFSYHSGL VATHAFGSPW | 1140 |
| WEWPFIARPI WFYSGQSLLP ADQVSSIVSM GNPAIWWPGT LAALVTAAWL VKGRSKGMLV | 1200 |
| VVVGFASQYI PWMLVPRLTF IYHYFAMVPF IILVLTYWFK RGMEANPRLN YAVYGYVGIC | 1260 |
| IALFAWFYPI LSGMTISKTY ASLLKWFQSW HFYS | 1294 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 869 | 1102 | 5.5e-52 | 0.9147982062780269 |
| CBM16 | 38 | 157 | 5.3e-18 | 0.9224137931034483 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5542 | COG5542 | 5.34e-26 | 258 | 614 | 42 | 369 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| 406576 | pfam16192 | 1.28e-23 | 1114 | 1287 | 1 | 197 | PMT_4TMC C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 1.87e-22 | 874 | 1293 | 50 | 696 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG4346 | COG4346 | 5.77e-15 | 884 | 1235 | 122 | 405 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam13231 | PMT_2 | 1.41e-13 | 897 | 1035 | 2 | 129 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSF45552.1 | 0.0 | 35 | 1292 | 27 | 1273 |
| CQR52353.1 | 0.0 | 32 | 1292 | 24 | 1275 |
| QUL55595.1 | 0.0 | 35 | 1292 | 27 | 1278 |
| AIQ39188.1 | 0.0 | 35 | 1292 | 27 | 1279 |
| ASA25569.1 | 0.0 | 35 | 1292 | 27 | 1268 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6P25_B | 8.76e-06 | 1141 | 1290 | 581 | 751 | Structureof S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor and a peptide acceptor [Saccharomyces cerevisiae W303],6P2R_B Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor [Saccharomyces cerevisiae W303] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WN04 | 7.54e-30 | 874 | 1290 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 7.54e-30 | 874 | 1290 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| L8F4Z2 | 7.23e-29 | 866 | 1290 | 59 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| Q8NRZ6 | 1.04e-26 | 896 | 1290 | 98 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
| Q5ACU3 | 4.59e-07 | 1114 | 1290 | 516 | 716 | Dolichyl-phosphate-mannose--protein mannosyltransferase 5 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
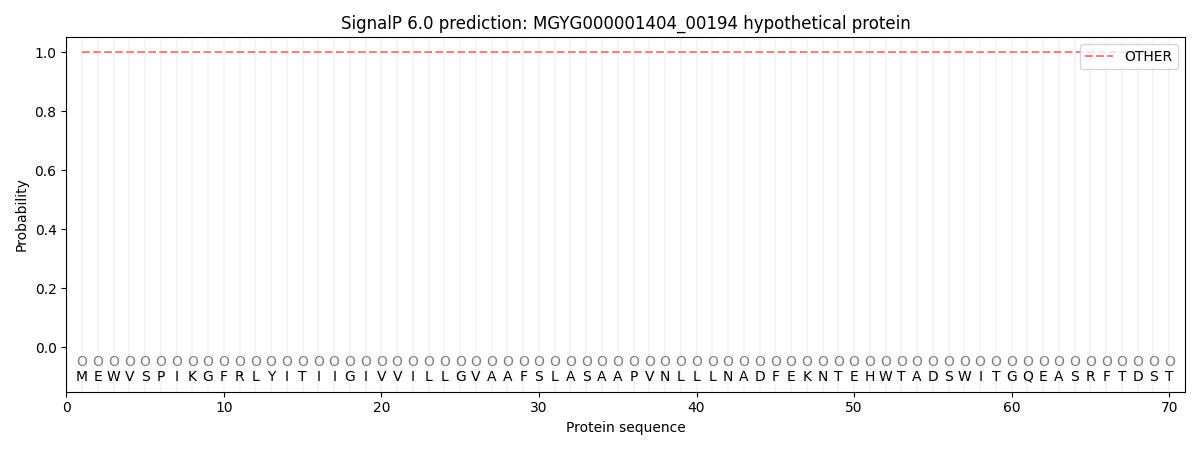
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999935 | 0.000086 | 0.000001 | 0.000001 | 0.000000 | 0.000011 |
TMHMM Annotations download full data without filtering help
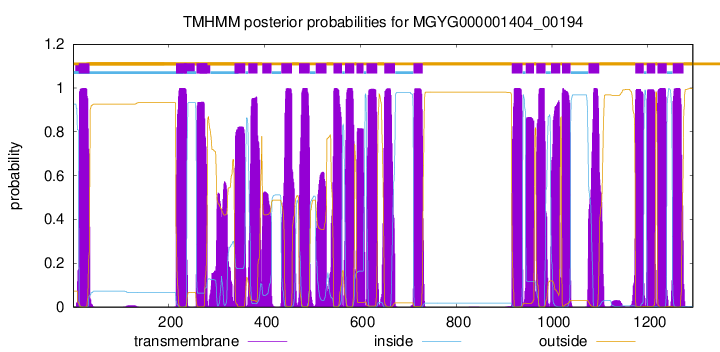
| start | end |
|---|---|
| 12 | 34 |
| 215 | 237 |
| 258 | 280 |
| 337 | 359 |
| 366 | 385 |
| 395 | 414 |
| 435 | 457 |
| 472 | 494 |
| 507 | 529 |
| 544 | 561 |
| 568 | 587 |
| 592 | 606 |
| 613 | 635 |
| 650 | 672 |
| 711 | 730 |
| 916 | 938 |
| 945 | 962 |
| 967 | 986 |
| 998 | 1017 |
| 1021 | 1038 |
| 1076 | 1098 |
| 1174 | 1191 |
| 1198 | 1215 |
| 1220 | 1239 |
| 1252 | 1274 |
