You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001407_02333
You are here: Home > Sequence: MGYG000001407_02333
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_J senegalense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_J; Clostridium_J senegalense | |||||||||||
| CAZyme ID | MGYG000001407_02333 | |||||||||||
| CAZy Family | CBM12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 119979; End: 122429 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03296 | M6dom_TIGR03296 | 7.42e-47 | 150 | 429 | 1 | 285 | M6 family metalloprotease domain. This model describes a metalloproteinase domain, with a characteristic HExxH motif. Examples of this domain are found in proteins in the family of immune inhibitor A, which cleaves antibacterial peptides, and in other, only distantly related proteases. This model is built to be broader and more inclusive than pfam05547. |
| pfam03272 | Mucin_bdg | 2.62e-28 | 528 | 642 | 1 | 116 | Putative mucin or carbohydrate-binding module. This family is the putative binding domain for the substrates of enhancin, and other similar metallopeptidases. This is not the enzymically active, peptidase, part of the proteins - see pfam13402. |
| cd12214 | ChiA1_BD | 1.35e-15 | 771 | 813 | 1 | 43 | chitin-binding domain of Chi A1-like proteins. This group contains proteins related to the chitin binding domain of chitinase A1 (ChiA1) of Bacillus circulans WL-12. Glycosidase ChiA1 hydrolyzes chitin and is comprised of several domains: the C-terminal chitin binding domain, an N-terminal and catalytic domain, and 2 fibronectin type III-like domains. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. Bacillus circulans WL-12 ChiA1 facilitates invasion of fungal cell walls. The ChiAi chitin binding domain is required for the specific recognition of insoluble chitin. although topologically and structurally related, ChiA1 lacks the characteristic aromatic residues of Erwinia chrysanthemi endoglucanase Z (CBD(EGZ)). |
| pfam05547 | Peptidase_M6 | 6.54e-11 | 172 | 439 | 60 | 350 | Immune inhibitor A peptidase M6. The insect pathogenic Gram-positive Bacillus thuringiensis secretes immune inhibitor A, a metallopeptidase, which specifically cleaves host antibacterial proteins. A homolog of immune inhibitor A, PrtV, has been identified in the Gram-negative human pathogen Vibrio cholerae. |
| COG4412 | COG4412 | 2.35e-08 | 173 | 421 | 186 | 473 | Bacillopeptidase F, M6 metalloprotease family [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRI51831.1 | 0.0 | 1 | 816 | 1 | 683 |
| APH17289.1 | 0.0 | 1 | 816 | 1 | 682 |
| ACA55272.1 | 0.0 | 1 | 816 | 1 | 682 |
| AWB30616.1 | 0.0 | 1 | 816 | 1 | 682 |
| ACO86706.1 | 0.0 | 1 | 816 | 1 | 682 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ILV_A | 3.24e-09 | 764 | 812 | 197 | 245 | Structureof the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4ILV_B Structure of the dioxygenase domain of SACTE_2871, a novel dioxygenase carbohydrate-binding protein fusion from the cellulolytic bacterium Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 1ED7_A | 4.71e-09 | 772 | 814 | 2 | 44 | ChainA, CHITINASE A1 [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 9.28e-09 | 730 | 814 | 616 | 698 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| P27050 | 1.58e-07 | 767 | 815 | 29 | 77 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
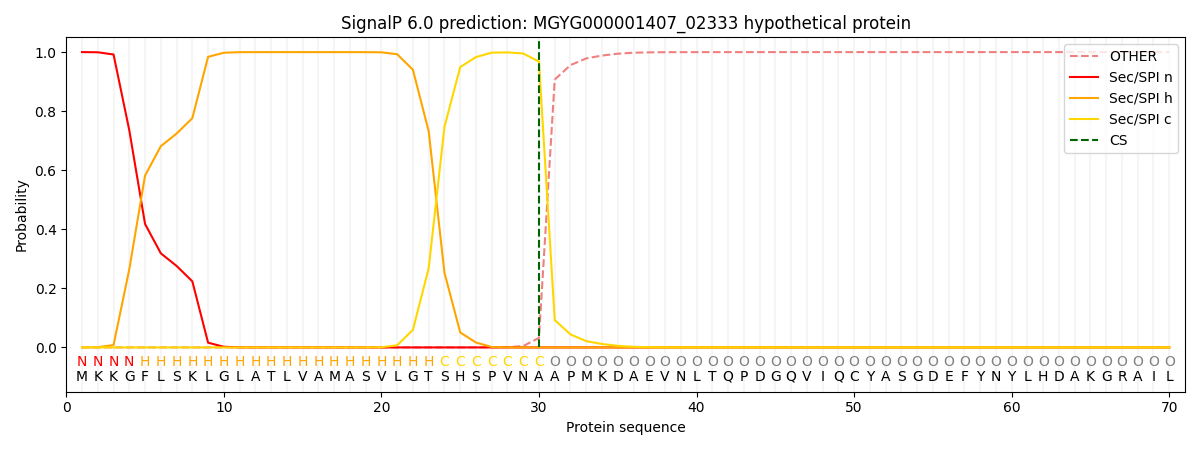
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000339 | 0.998851 | 0.000210 | 0.000248 | 0.000187 | 0.000155 |
