You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001407_03570
You are here: Home > Sequence: MGYG000001407_03570
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_J senegalense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_J; Clostridium_J senegalense | |||||||||||
| CAZyme ID | MGYG000001407_03570 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 226045; End: 227592 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06543 | GH18_PF-ChiA-like | 3.34e-120 | 58 | 363 | 1 | 294 | PF-ChiA is an uncharacterized chitinase found in the hyperthermophilic archaeon Pyrococcus furiosus with a glycosyl hydrolase family 18 (GH18) catalytic domain as well as a cellulose-binding domain. Members of this domain family are found not only in archaea but also in eukaryotes and prokaryotes. PF-ChiA exhibits hydrolytic activity toward both colloidal and crystalline (beta/alpha) chitins at high temperature. |
| cd12215 | ChiC_BD | 3.28e-12 | 467 | 508 | 1 | 41 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
| cd12215 | ChiC_BD | 1.13e-10 | 393 | 438 | 2 | 42 | Chitin-binding domain of chitinase C. Chitin-binding domain of chitinase C (ChiC) of Streptomyces griseus and related proteins. Chitinase C is a family 19 chitinase, and consists of a N-terminal chitin binding domain and a C-terminal chitin-catalytic domain that effects degradation. Chitinases function in invertebrates in the degradation of old exoskeletons, in fungi to utilize chitin in cell walls, and in bacteria which use chitin as an energy source. ChiC contains the characteristic chitin-binding aromatic residues. |
| smart00495 | ChtBD3 | 3.02e-10 | 466 | 507 | 1 | 41 | Chitin-binding domain type 3. |
| COG3979 | COG3979 | 4.83e-09 | 391 | 511 | 40 | 145 | Chitodextrinase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APF27458.1 | 7.18e-289 | 1 | 514 | 1 | 526 |
| AVQ43846.1 | 4.14e-288 | 1 | 514 | 1 | 526 |
| AVQ40292.1 | 4.14e-288 | 1 | 514 | 1 | 526 |
| AKC61813.1 | 1.68e-287 | 1 | 514 | 1 | 526 |
| AVP59775.1 | 1.68e-287 | 1 | 514 | 1 | 526 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2DSK_A | 2.16e-34 | 60 | 342 | 12 | 272 | Crystalstructure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus],2DSK_B Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
| 3A4W_A | 1.48e-33 | 60 | 342 | 12 | 272 | Crystalstructures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4W_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus] |
| 3A4X_A | 2.81e-33 | 60 | 342 | 12 | 272 | Crystalstructures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3A4X_B Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides [Pyrococcus furiosus],3AFB_A Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus],3AFB_B Crystal structures of catalytic site mutants of active domain 2 of chitinase from Pyrococcus furiosus [Pyrococcus furiosus] |
| 2RTS_A | 8.04e-06 | 467 | 508 | 25 | 65 | Chitinbinding domain1 [Pyrococcus furiosus DSM 3638] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P13656 | 6.84e-52 | 42 | 365 | 571 | 894 | Probable bifunctional chitinase/lysozyme OS=Escherichia coli (strain K12) OX=83333 GN=chiA PE=1 SV=2 |
| O74199 | 1.43e-50 | 42 | 333 | 220 | 509 | Endochitinase 11 OS=Metarhizium anisopliae OX=5530 GN=chi11 PE=1 SV=1 |
| P14529 | 6.26e-19 | 73 | 367 | 56 | 326 | Chitinase OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=chiA2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
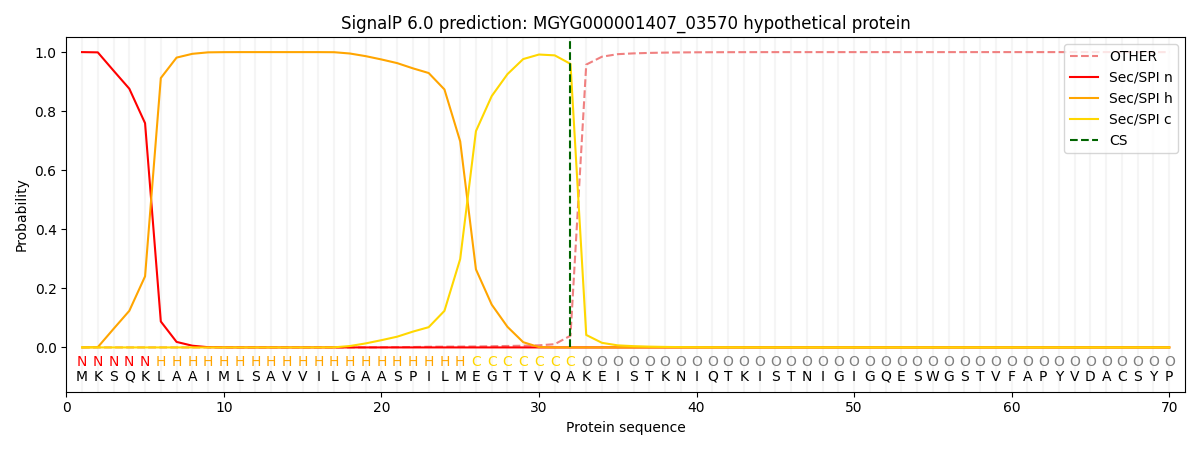
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000276 | 0.999081 | 0.000177 | 0.000167 | 0.000140 | 0.000130 |
