You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001415_02371
You are here: Home > Sequence: MGYG000001415_02371
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes communis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes communis | |||||||||||
| CAZyme ID | MGYG000001415_02371 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 702784; End: 704262 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH123 | 51 | 455 | 7.6e-143 | 0.7546468401486989 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL03317.1 | 0.0 | 1 | 468 | 1 | 468 |
| BBL07500.1 | 5.46e-314 | 1 | 468 | 1 | 468 |
| SCM55400.1 | 1.15e-189 | 19 | 468 | 24 | 474 |
| BBD46967.1 | 1.98e-187 | 19 | 455 | 32 | 469 |
| SCD18916.1 | 1.15e-186 | 16 | 468 | 21 | 474 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5L7V_A | 8.18e-109 | 32 | 468 | 6 | 446 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 1.25e-108 | 32 | 468 | 21 | 461 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5FQE_A | 4.96e-51 | 83 | 454 | 62 | 451 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQE_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FR0_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5FQG_A | 6.58e-50 | 83 | 454 | 62 | 451 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQH_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
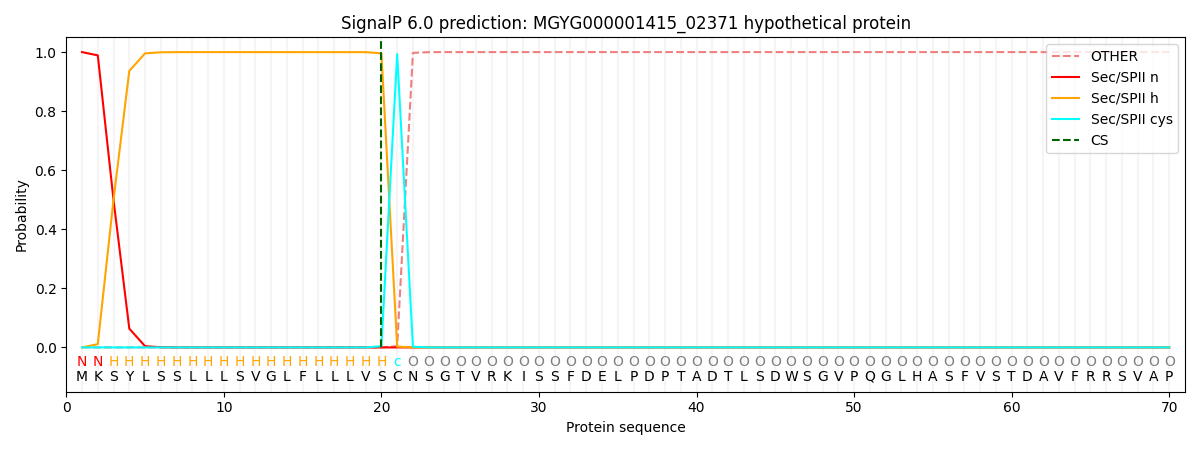
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000025 | 0.000000 | 0.000000 | 0.000000 |
