You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_00233
You are here: Home > Sequence: MGYG000001416_00233
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
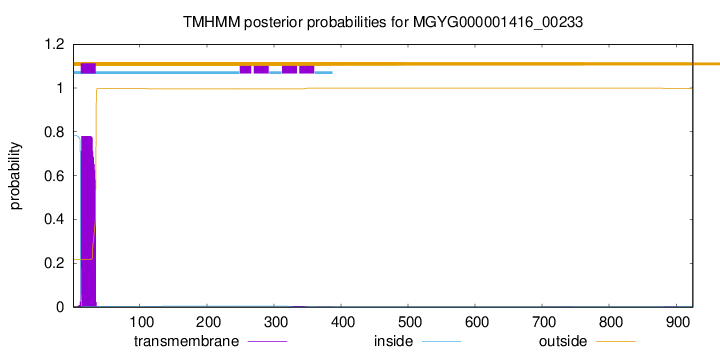
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_00233 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 267190; End: 269967 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 220 | 414 | 5.6e-25 | 0.8678414096916299 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 1.22e-27 | 433 | 829 | 18 | 387 | Predicted peptidase [General function prediction only]. |
| COG2382 | Fes | 3.31e-20 | 194 | 415 | 60 | 281 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam18435 | EstA_Ig_like | 2.01e-19 | 442 | 571 | 5 | 120 | Esterase Ig-like N-terminal domain. This is an N-terminal immunoglobulin (Ig)-like domain found in esterases such as EstA. Analysis of the EstA structure confirms that it is a member of the alpha/beta hydrolase family, with a conserved Ser-Asp-His catalytic triad. The Ig-like domain presumably plays a role in the multimerization of EstA into an unusual hexameric structure. Additionally, it may also participate in the catalysis of EstA by guiding the substrate to the active site. |
| pfam00756 | Esterase | 5.12e-11 | 221 | 414 | 12 | 231 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| COG1506 | DAP2 | 2.65e-07 | 569 | 733 | 340 | 506 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ59636.1 | 4.16e-222 | 4 | 436 | 5 | 437 |
| QJW36208.1 | 2.73e-31 | 422 | 925 | 669 | 1183 |
| QMW66742.1 | 3.04e-25 | 17 | 381 | 14 | 393 |
| QIA09957.1 | 2.45e-21 | 106 | 352 | 47 | 269 |
| ADQ79787.1 | 8.58e-20 | 106 | 352 | 69 | 293 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 6.91e-22 | 436 | 829 | 6 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 1JJF_A | 4.46e-20 | 195 | 356 | 24 | 181 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 1.10e-19 | 195 | 356 | 24 | 181 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
| 3WYD_A | 3.52e-13 | 606 | 772 | 20 | 178 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
| 7B5V_A | 5.11e-13 | 201 | 353 | 137 | 297 | ChainA, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B5V_B Chain B, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B6B_A Chain A, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836],7B6B_B Chain B, Carbohydrate Esterase family 1 protein with an N-terminal carbohydrate binding module family 48 [Dysgonomonas mossii DSM 22836] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 8.50e-18 | 195 | 356 | 43 | 200 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| P31471 | 1.61e-12 | 104 | 367 | 73 | 317 | Uncharacterized protein YieL OS=Escherichia coli (strain K12) OX=83333 GN=yieL PE=4 SV=3 |
| D5EY13 | 2.73e-11 | 195 | 353 | 492 | 654 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| D5EXZ4 | 1.58e-09 | 201 | 353 | 429 | 588 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
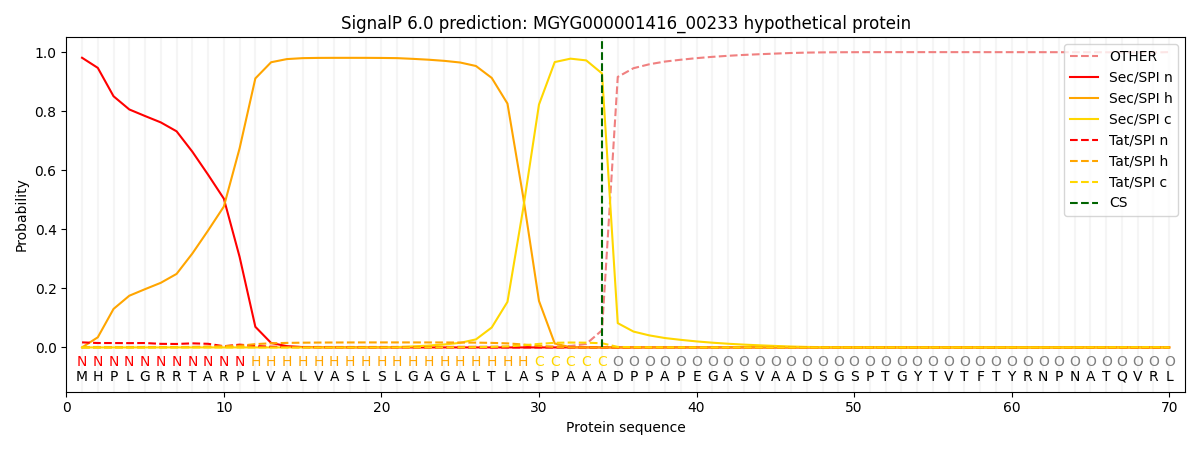
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002893 | 0.977158 | 0.000959 | 0.018279 | 0.000411 | 0.000278 |