You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_00342
You are here: Home > Sequence: MGYG000001416_00342
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
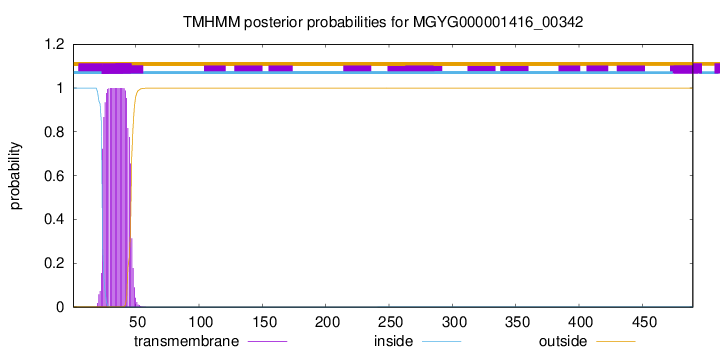
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_00342 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 376026; End: 377498 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 365 | 488 | 9.6e-31 | 0.6702127659574468 |
| GH16 | 58 | 287 | 7.4e-18 | 0.9682539682539683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00652 | Ricin_B_lectin | 6.75e-26 | 367 | 487 | 4 | 126 | Ricin-type beta-trefoil lectin domain. |
| NF035929 | lectin_1 | 1.20e-24 | 374 | 489 | 721 | 836 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
| cd00161 | RICIN | 1.27e-23 | 370 | 489 | 6 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
| smart00458 | RICIN | 1.92e-23 | 369 | 490 | 2 | 118 | Ricin-type beta-trefoil. Carbohydrate-binding domain formed from presumed gene triplication. |
| cd00413 | Glyco_hydrolase_16 | 1.23e-22 | 59 | 287 | 1 | 207 | glycosyl hydrolase family 16. The O-Glycosyl hydrolases are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycosyl hydrolase family 16. Family 16 includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGZ39149.1 | 3.70e-138 | 42 | 459 | 25 | 424 |
| ADB29204.1 | 3.68e-113 | 51 | 318 | 42 | 314 |
| AGL20089.1 | 1.41e-108 | 28 | 328 | 7 | 306 |
| QZD57928.1 | 1.43e-108 | 47 | 341 | 3 | 296 |
| UAC01072.1 | 4.69e-108 | 44 | 403 | 33 | 357 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
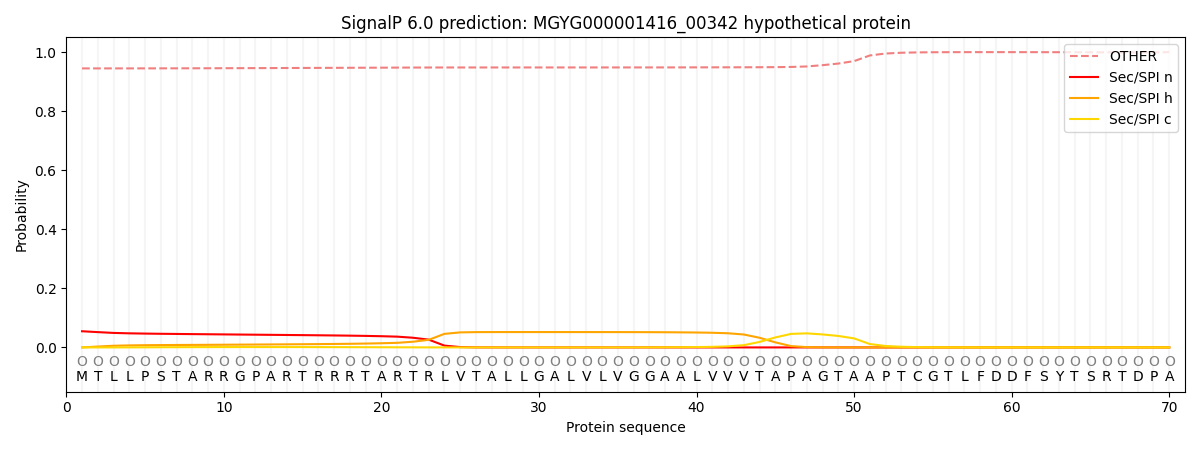
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.947799 | 0.051038 | 0.000294 | 0.000172 | 0.000119 | 0.000608 |