You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_00373
You are here: Home > Sequence: MGYG000001416_00373
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
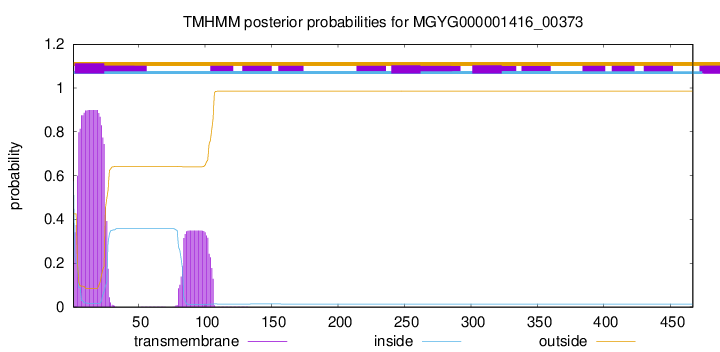
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_00373 | |||||||||||
| CAZy Family | CBM3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 413860; End: 415263 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM3 | 31 | 113 | 3.2e-18 | 0.9886363636363636 |
| GH16 | 204 | 433 | 6e-18 | 0.9682539682539683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00413 | Glyco_hydrolase_16 | 5.80e-24 | 205 | 435 | 1 | 209 | glycosyl hydrolase family 16. The O-Glycosyl hydrolases are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A glycosyl hydrolase classification system based on sequence similarity has led to the definition of more than 95 different families inlcuding glycosyl hydrolase family 16. Family 16 includes lichenase, xyloglucan endotransglycosylase (XET), beta-agarase, kappa-carrageenase, endo-beta-1,3-glucanase, endo-beta-1,3-1,4-glucanase, and endo-beta-galactosidase, all of which have a conserved jelly roll fold with a deep active site channel harboring the catalytic residues. |
| pfam00942 | CBM_3 | 1.32e-20 | 30 | 112 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 5.49e-15 | 30 | 113 | 1 | 83 | Cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACZ91718.1 | 4.42e-251 | 15 | 467 | 19 | 467 |
| AWS44283.1 | 1.09e-223 | 22 | 467 | 24 | 468 |
| ANZ41235.1 | 2.06e-185 | 21 | 464 | 24 | 458 |
| QFZ19899.1 | 9.63e-184 | 21 | 464 | 24 | 458 |
| QQQ80609.1 | 2.56e-183 | 8 | 466 | 7 | 455 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2L8A_A | 2.25e-33 | 27 | 174 | 3 | 148 | Structureof a novel CBM3 lacking the calcium-binding site [Bacillus subtilis],6UFV_A Crystal structure of the CBM3 from Bacillus subtilis at 1.06 angstrom resolution [Bacillus subtilis subsp. subtilis str. 168],6UFW_A Crystal structure of the CBM3 from Bacillus subtilis at 1.28 angstrom resolution [Bacillus subtilis subsp. subtilis str. 168] |
| 6D5B_A | 2.96e-32 | 26 | 177 | 17 | 170 | Structureof Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_B Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_C Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_D Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_E Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_F Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_G Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_H Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_I Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_J Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_K Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_L Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 4JO5_A | 8.83e-28 | 26 | 177 | 17 | 175 | ChainA, Cellulosome anchoring protein cohesin region [Acetivibrio thermocellus YS] |
| 1NBC_A | 2.28e-26 | 28 | 174 | 2 | 155 | ChainA, Cellulosomal Scaffolding Protein A [Acetivibrio thermocellus],1NBC_B Chain B, Cellulosomal Scaffolding Protein A [Acetivibrio thermocellus] |
| 4B9F_A | 1.05e-25 | 28 | 171 | 2 | 152 | ChainA, Cellulosomal-scaffolding Protein A [Acetivibrio thermocellus ATCC 27405],4B9F_B Chain B, Cellulosomal-scaffolding Protein A [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10475 | 1.77e-29 | 28 | 174 | 354 | 498 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P22533 | 2.57e-29 | 26 | 180 | 568 | 724 | Beta-mannanase/endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=manA PE=1 SV=2 |
| P10474 | 3.09e-29 | 26 | 179 | 420 | 575 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| P22534 | 1.21e-28 | 26 | 179 | 705 | 860 | Endoglucanase A OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celA PE=3 SV=2 |
| P23549 | 1.53e-28 | 28 | 174 | 354 | 498 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
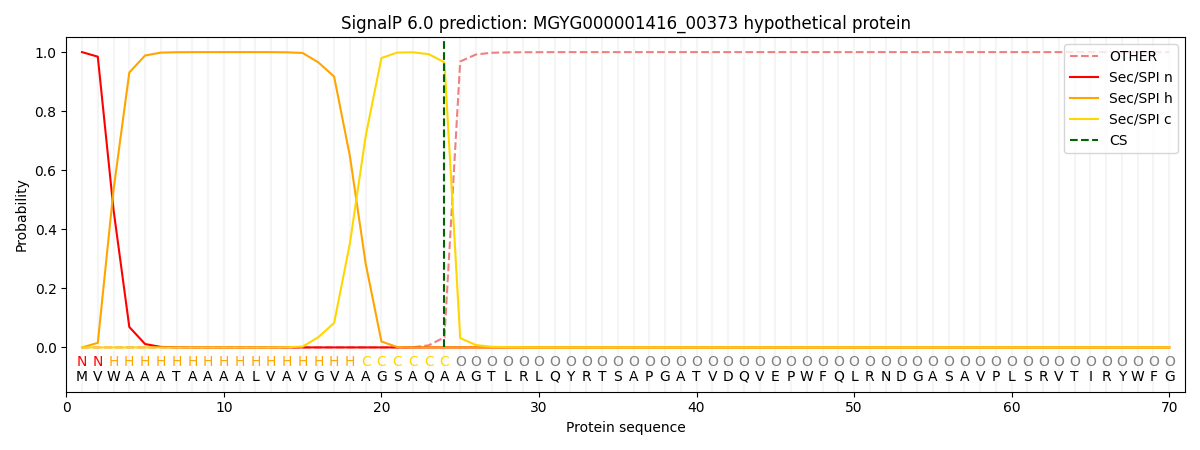
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000547 | 0.998514 | 0.000214 | 0.000273 | 0.000221 | 0.000209 |