You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_00423
You are here: Home > Sequence: MGYG000001416_00423
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
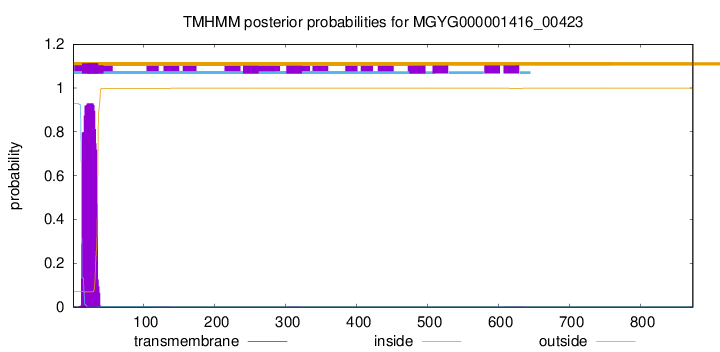
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_00423 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 465316; End: 467940 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 566 | 846 | 6.6e-71 | 0.4594594594594595 |
| CBM32 | 198 | 311 | 1.3e-27 | 0.9274193548387096 |
| CBM32 | 47 | 160 | 2.2e-26 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 7.18e-22 | 196 | 309 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 4.65e-20 | 48 | 159 | 3 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.71e-06 | 197 | 298 | 14 | 130 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 9.21e-06 | 190 | 310 | 3 | 136 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| cd00057 | FA58C | 2.92e-04 | 48 | 120 | 15 | 92 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARU52139.1 | 0.0 | 15 | 872 | 20 | 871 |
| QJW37750.1 | 0.0 | 38 | 872 | 40 | 877 |
| QLQ35640.1 | 0.0 | 6 | 872 | 7 | 856 |
| ATO13362.1 | 0.0 | 5 | 872 | 12 | 862 |
| SBT42009.1 | 0.0 | 2 | 872 | 8 | 861 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4TZ1_A | 3.34e-244 | 323 | 872 | 2 | 548 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4TZ3_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4TZ5_A Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4TZ5_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4TYV_A | 3.59e-244 | 323 | 872 | 4 | 550 | Ensemblerefinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4TYV_B Ensemble refinement of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E] |
| 4PEX_A | 5.14e-244 | 323 | 872 | 14 | 560 | Structureof the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEX_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with glucose [Streptomyces sp. SirexAA-E],4PEY_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritriose [Streptomyces sp. SirexAA-E],4PEZ_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminaritetraose [Streptomyces sp. SirexAA-E],4PF0_A Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E],4PF0_B Structure of the E502A variant of sacteLam55A from Streptomyces sp. SirexAA-E in complex with laminarihexaose [Streptomyces sp. SirexAA-E] |
| 4PEW_A | 8.32e-243 | 323 | 872 | 14 | 560 | Structureof sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E],4PEW_B Structure of sacteLam55A from Streptomyces sp. SirexAA-E [Streptomyces sp. SirexAA-E] |
| 2RV9_A | 4.44e-28 | 190 | 314 | 7 | 136 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2NFJ9 | 1.68e-243 | 323 | 872 | 58 | 604 | Exo-beta-1,3-glucanase OS=Streptomyces sp. (strain SirexAA-E / ActE) OX=862751 GN=SACTE_4363 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
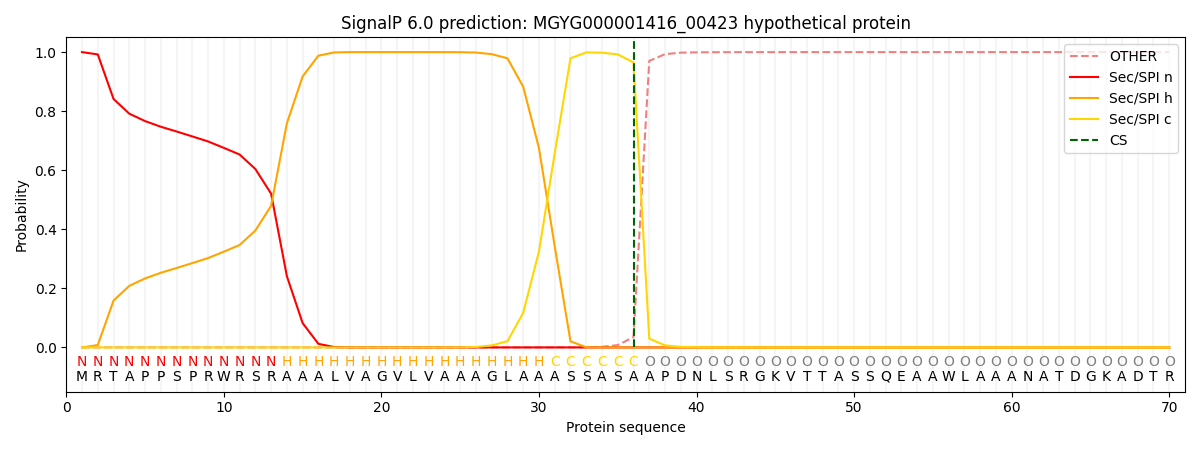
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000355 | 0.998914 | 0.000187 | 0.000213 | 0.000172 | 0.000154 |