You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_00516
You are here: Home > Sequence: MGYG000001416_00516
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
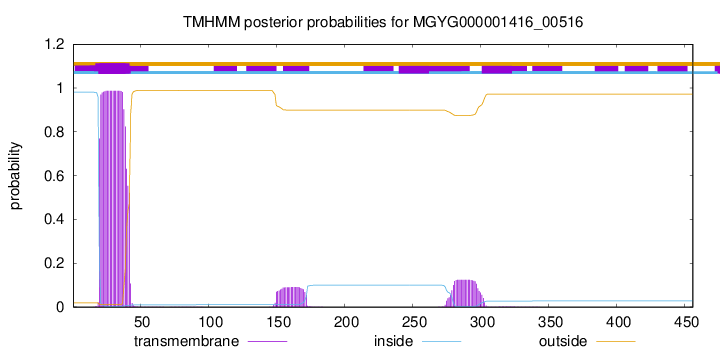
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_00516 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 556263; End: 557633 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 58 | 318 | 5.4e-56 | 0.991304347826087 |
| CBM13 | 338 | 455 | 3.9e-27 | 0.6382978723404256 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02182 | GH16_Strep_laminarinase_like | 1.78e-117 | 52 | 319 | 1 | 259 | Streptomyces laminarinase-like, member of glycosyl hydrolase family 16. Proteins similar to Streptomyces sioyaensis beta-1,3-glucanase (laminarinase) present in Actinomycetales as well as Peziomycotina. Laminarinases belong to glycosyl hydrolase family 16 and hydrolyze the glycosidic bond of the 1,3-beta-linked glucan, a major component of fungal and plant cell walls and the structural and storage polysaccharides (laminarin) of marine macro-algae. Members of the GH16 family have a conserved jelly roll fold with an active site channel. |
| cd08023 | GH16_laminarinase_like | 8.08e-49 | 57 | 318 | 1 | 235 | Laminarinase, member of the glycosyl hydrolase family 16. Laminarinase, also known as glucan endo-1,3-beta-D-glucosidase, is a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| NF035929 | lectin_1 | 4.70e-33 | 339 | 455 | 720 | 836 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
| pfam00652 | Ricin_B_lectin | 5.77e-21 | 339 | 453 | 10 | 126 | Ricin-type beta-trefoil lectin domain. |
| cd00161 | RICIN | 7.95e-21 | 335 | 455 | 5 | 124 | Ricin-type beta-trefoil; Carbohydrate-binding domain formed from presumed gene triplication. The domain is found in a variety of molecules serving diverse functions such as enzymatic activity, inhibitory toxicity and signal transduction. Highly specific ligand binding occurs on exposed surfaces of the compact domain sturcture. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDT77088.1 | 1.02e-162 | 42 | 324 | 27 | 309 |
| BBH64583.1 | 1.39e-162 | 39 | 334 | 23 | 320 |
| BCJ42700.1 | 7.92e-161 | 31 | 329 | 5 | 305 |
| BAL92475.1 | 7.69e-159 | 51 | 376 | 33 | 366 |
| SLM04226.1 | 8.17e-159 | 37 | 327 | 22 | 312 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DGT_A | 3.31e-126 | 45 | 315 | 1 | 271 | The1.5 A crystal structure of endo-1,3-beta-glucanase from Streptomyces sioyaensis [Streptomyces sioyaensis] |
| 7EO3_A | 4.86e-103 | 51 | 322 | 8 | 282 | ChainA, 1,3-beta-glucanase [Actinomycetia bacterium] |
| 3ATG_A | 9.56e-37 | 56 | 327 | 6 | 247 | endo-1,3-beta-glucanasefrom Cellulosimicrobium cellulans [Cellulosimicrobium cellulans] |
| 2HYK_A | 2.70e-34 | 54 | 321 | 8 | 245 | Thecrystal structure of an endo-beta-1,3-glucanase from alkaliphilic Nocardiopsis sp.strain F96 [Nocardiopsis sp. F96] |
| 2VY0_A | 7.77e-31 | 53 | 318 | 13 | 259 | TheX-ray structure of endo-beta-1,3-glucanase from Pyrococcus furiosus [Pyrococcus furiosus],2VY0_B The X-ray structure of endo-beta-1,3-glucanase from Pyrococcus furiosus [Pyrococcus furiosus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C1IE32 | 3.54e-19 | 53 | 300 | 20 | 245 | Glucan endo-1,3-beta-glucosidase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
| P23903 | 1.98e-18 | 52 | 318 | 422 | 679 | Glucan endo-1,3-beta-glucosidase A1 OS=Niallia circulans OX=1397 GN=glcA PE=1 SV=1 |
| P45798 | 3.94e-18 | 53 | 318 | 40 | 282 | Beta-glucanase OS=Rhodothermus marinus OX=29549 GN=bglA PE=1 SV=1 |
| Q9ZG90 | 1.37e-12 | 56 | 318 | 59 | 287 | Keratan-sulfate endo-1,4-beta-galactosidase OS=Sphingobacterium multivorum OX=28454 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002940 | 0.980546 | 0.000486 | 0.015434 | 0.000352 | 0.000233 |