You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_01656
You are here: Home > Sequence: MGYG000001416_01656
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
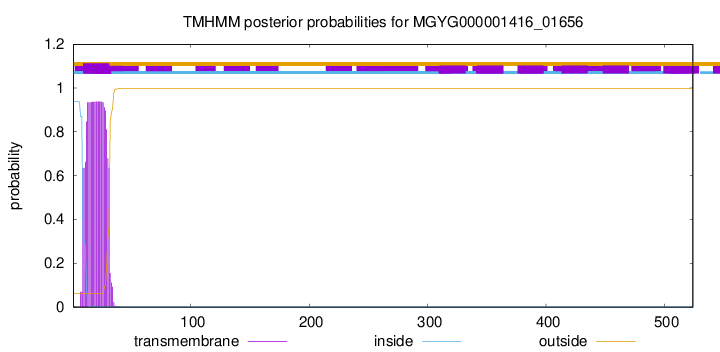
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_01656 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | Endoglucanase E-2 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1768999; End: 1770573 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 60 | 381 | 3.6e-77 | 0.9455782312925171 |
| CBM2 | 423 | 523 | 3.1e-16 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01341 | Glyco_hydro_6 | 4.39e-64 | 75 | 387 | 11 | 292 | Glycosyl hydrolases family 6. |
| COG5297 | CelA1 | 6.18e-25 | 89 | 500 | 165 | 538 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| pfam00553 | CBM_2 | 1.42e-09 | 423 | 523 | 1 | 101 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 2.03e-08 | 434 | 504 | 5 | 75 | CBD_II domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALK04194.1 | 4.68e-173 | 2 | 523 | 3 | 532 |
| ARU50913.1 | 2.58e-172 | 1 | 523 | 1 | 531 |
| QEO14244.1 | 1.31e-158 | 8 | 523 | 8 | 518 |
| QNO37390.1 | 2.29e-158 | 49 | 523 | 39 | 513 |
| QIG39320.1 | 3.16e-131 | 4 | 523 | 5 | 524 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1UOZ_A | 2.47e-33 | 39 | 414 | 21 | 313 | Structureof the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with thiocellopentaose at 1.1 angstrom [Mycobacterium tuberculosis H37Rv] |
| 1UP0_A | 7.69e-33 | 43 | 414 | 4 | 292 | Structureof the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with cellobiose at 1.75 angstrom [Mycobacterium tuberculosis H37Rv] |
| 1UP3_A | 7.69e-33 | 43 | 414 | 4 | 292 | Structureof the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with METHYL-CELLOBIOSYL-4-DEOXY-4-THIO-BETA-D-CELLOBIOSIDE at 1.6 angstrom [Mycobacterium tuberculosis H37Rv] |
| 1UP2_A | 1.46e-32 | 43 | 414 | 4 | 292 | Structureof the endoglucanase Cel6 from Mycobacterium tuberculosis in complex with glucose-isofagomine at 1.9 angstrom [Mycobacterium tuberculosis H37Rv] |
| 1TML_A | 6.41e-26 | 75 | 384 | 33 | 271 | CRYSTALSTRUCTURE OF THE CATALYTIC DOMAIN OF A THERMOPHILIC ENDOCELLULASE [Thermobifida fusca],2BOD_X Catalytic domain of endo-1,4-glucanase Cel6A from Thermobifida fusca in complex with methyl cellobiosyl-4-thio-beta-cellobioside [Thermobifida fusca] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26414 | 1.43e-36 | 5 | 509 | 2 | 446 | Endoglucanase A OS=Thermobispora bispora OX=2006 GN=celA PE=3 SV=1 |
| P26222 | 1.81e-29 | 75 | 515 | 64 | 436 | Endoglucanase E-2 OS=Thermobifida fusca OX=2021 GN=celB PE=1 SV=2 |
| P33682 | 1.05e-25 | 49 | 411 | 37 | 318 | Endoglucanase 1 OS=Streptomyces halstedii OX=1944 GN=celA1 PE=1 SV=2 |
| P13933 | 7.83e-24 | 75 | 362 | 107 | 324 | Endoglucanase 1 OS=Streptomyces sp. (strain KSM-9) OX=74575 GN=casA PE=1 SV=3 |
| P07984 | 5.47e-23 | 43 | 414 | 168 | 447 | Endoglucanase A OS=Cellulomonas fimi OX=1708 GN=cenA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
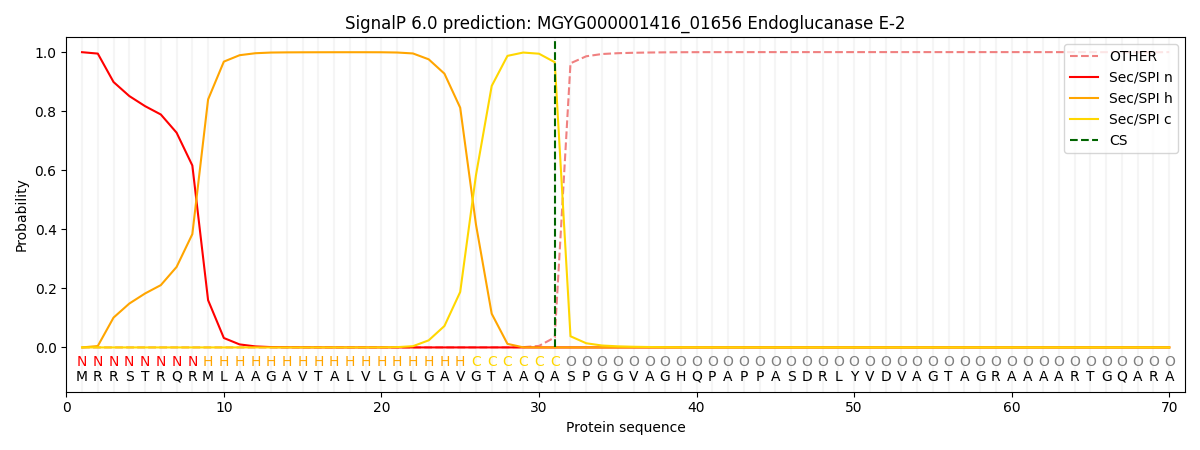
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000653 | 0.997723 | 0.000289 | 0.000830 | 0.000273 | 0.000196 |