You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001416_01683
You are here: Home > Sequence: MGYG000001416_01683
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cellulomonas massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas massiliensis | |||||||||||
| CAZyme ID | MGYG000001416_01683 | |||||||||||
| CAZy Family | CBM12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1804985; End: 1806478 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd21112 | alphaLP-like | 4.16e-66 | 204 | 377 | 1 | 188 | alpha-lytic protease (alpha-LP), a bacterial serine protease of the chymotrypsin family, and similar proteins. This family represents the catalytic domain of alpha-lytic protease (alpha-LP) and its closely-related homologs. Alpha-lytic protease (EC 3.4.21.12; also called alpha-lytic endopeptidase), originally isolated from the myxobacterium Lysobacter enzymogenes, belongs to the MEROPS peptidase family S1, subfamily S1E (streptogrisin A subfamily). It is synthesized as a pro-enzyme, thus having two domains; the N-terminal pro-domain acts as a foldase, required transiently for the correct folding of the protease domain, and also acts as a potent inhibitor of the mature enzyme, while the C-terminal domain catalyzes the cleavage of peptide bonds. Members of the alpha-lytic protease subfamily include Nocardiopsis alba protease (NAPase), a secreted chymotrypsin from the alkaliphile Cellulomonas bogoriensis, streptogrisins (SPG-A, SPG-B, SPG-C, and SPG-D), and Thermobifida fusca protease A (TFPA). These serine proteases have characteristic kinetic stability, exhibited by their extremely slow unfolding kinetics. The active site, characteristic of serine proteases, contains the catalytic triad consisting of serine acting as a nucleophile, aspartate as an electrophile, and histidine as a base, all required for activity. This model represents the C-terminal catalytic domain of alpha-lytic proteases. |
| pfam00089 | Trypsin | 1.43e-11 | 204 | 374 | 1 | 217 | Trypsin. |
| pfam02983 | Pro_Al_protease | 4.05e-05 | 131 | 165 | 2 | 36 | Alpha-lytic protease prodomain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ76543.1 | 7.80e-134 | 5 | 383 | 3 | 371 |
| ADB33067.1 | 4.57e-127 | 8 | 383 | 1 | 371 |
| CCH28016.1 | 2.86e-126 | 50 | 442 | 39 | 423 |
| AUI59717.1 | 1.48e-125 | 35 | 383 | 26 | 375 |
| QFZ16630.1 | 9.80e-125 | 3 | 437 | 2 | 419 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2EA3_A | 4.98e-90 | 203 | 383 | 2 | 183 | CrystalStructure Of Cellulomonas Bogoriensis Chymotrypsin [Cellulomonas bogoriensis] |
| 2PFE_A | 3.67e-70 | 204 | 382 | 3 | 185 | ChainA, Alkaline serine protease [Thermobifida fusca YX],2PFE_B Chain B, Alkaline serine protease [Thermobifida fusca YX] |
| 2OUA_A | 2.41e-56 | 203 | 382 | 2 | 188 | CrystalStructure of Nocardiopsis Protease (NAPase) [Nocardiopsis alba],2OUA_B Crystal Structure of Nocardiopsis Protease (NAPase) [Nocardiopsis alba] |
| 2SFA_A | 1.67e-47 | 204 | 381 | 1 | 191 | SerineProteinase From Streptomyces Fradiae Atcc 14544 [Streptomyces fradiae] |
| 1SGD_E | 1.52e-45 | 204 | 380 | 1 | 184 | ChainE, Streptogrisin B [Streptomyces griseus],1SGE_E Chain E, Streptogrisin B [Streptomyces griseus],1SGN_E Chain E, Streptogrisin B [Streptomyces griseus],1SGY_E Chain E, Streptogrisin B [Streptomyces griseus],2GKV_E Chain E, Streptogrisin B [Streptomyces griseus],2NU0_E Chain E, Streptogrisin B, Protease B [Streptomyces griseus],2NU1_E Chain E, Streptogrisin B [Streptomyces griseus],2NU2_E Chain E, Streptogrisin B, Protease B [Streptomyces griseus],2NU3_E Chain E, Streptogrisin B, Proteinase B [Streptomyces griseus],2NU4_E Chain E, Streptogrisin B, Proteinase B [Streptomyces griseus],2QA9_E Crystal structure of the second tetrahedral intermediates of SGPB at pH 4.2 [Streptomyces griseus],2QAA_A Crystal structure of the second tetrahedral intermediates of SGPB at pH 7.3 [Streptomyces griseus],2QAA_B Crystal structure of the second tetrahedral intermediates of SGPB at pH 7.3 [Streptomyces griseus],2SGD_E ASP 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 [Streptomyces griseus],2SGE_E GLU 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 10.7 [Streptomyces griseus],2SGF_E PHE 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B [Streptomyces griseus],2SGP_E Chain E, PROTEINASE B [Streptomyces griseus],2SGQ_E GLN 18 VARIANT OF TURKEY OVOMUCOID INHIBITOR THIRD DOMAIN COMPLEXED WITH STREPTOMYCES GRISEUS PROTEINASE B AT PH 6.5 [Streptomyces griseus],3SGQ_E Chain E, Streptogrisin B [Streptomyces griseus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P52320 | 2.27e-106 | 1 | 437 | 6 | 440 | Streptogrisin-C OS=Streptomyces griseus OX=1911 GN=sprC PE=3 SV=1 |
| P52321 | 1.27e-49 | 83 | 377 | 88 | 388 | Streptogrisin-D OS=Streptomyces griseus OX=1911 GN=sprD PE=1 SV=1 |
| P00778 | 7.62e-48 | 54 | 382 | 44 | 396 | Alpha-lytic protease OS=Lysobacter enzymogenes OX=69 GN=alpha-LP PE=1 SV=3 |
| P00777 | 5.09e-47 | 150 | 380 | 62 | 298 | Streptogrisin-B OS=Streptomyces griseus OX=1911 GN=sprB PE=1 SV=2 |
| Q05308 | 1.84e-46 | 51 | 383 | 63 | 397 | Serine protease 1 OS=Rarobacter faecitabidus OX=13243 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
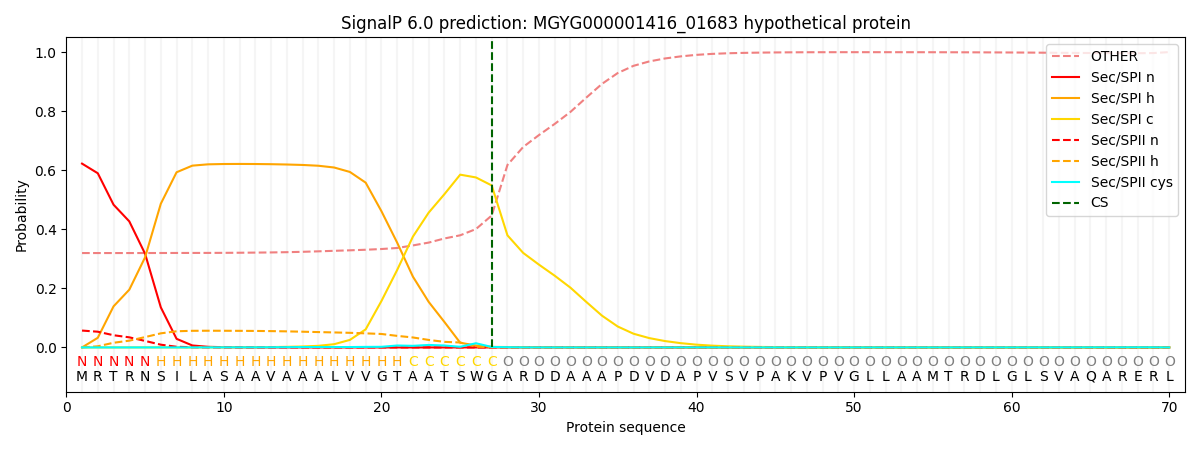
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.335130 | 0.601841 | 0.059026 | 0.002281 | 0.000951 | 0.000762 |

