You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001418_00197
You are here: Home > Sequence: MGYG000001418_00197
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Aeromicrobium massiliense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; Aeromicrobium massiliense | |||||||||||
| CAZyme ID | MGYG000001418_00197 | |||||||||||
| CAZy Family | GH114 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 201378; End: 202133 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH114 | 42 | 213 | 4e-48 | 0.8578947368421053 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03537 | Glyco_hydro_114 | 1.60e-61 | 25 | 238 | 3 | 219 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG3868 | COG3868 | 6.45e-12 | 44 | 197 | 94 | 248 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGG42551.1 | 1.38e-109 | 1 | 251 | 20 | 273 |
| SKB07859.1 | 3.36e-104 | 1 | 251 | 4 | 257 |
| AWB93365.1 | 5.72e-103 | 1 | 251 | 5 | 258 |
| QNL94108.1 | 6.98e-99 | 3 | 251 | 6 | 257 |
| ALX05635.1 | 1.09e-97 | 17 | 251 | 31 | 266 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
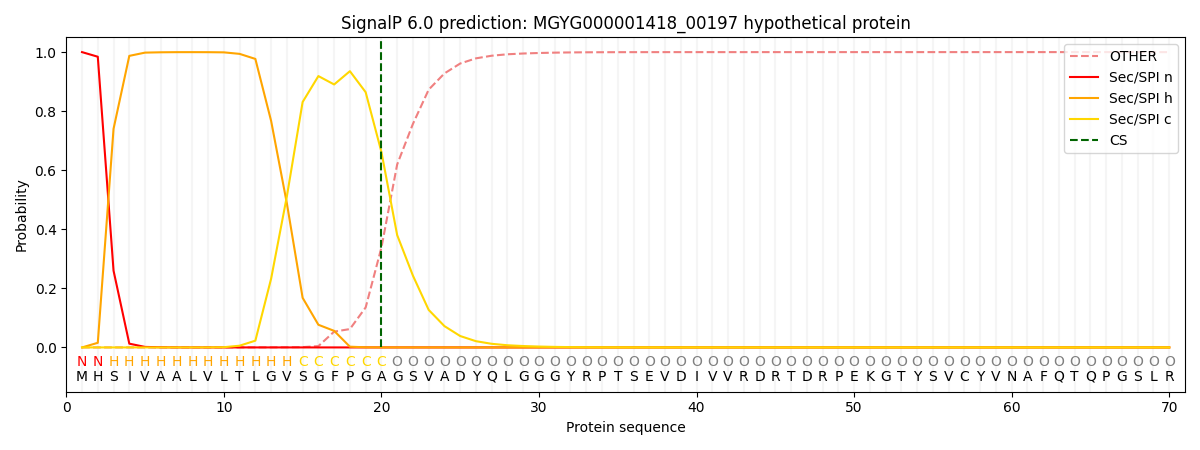
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000933 | 0.998117 | 0.000309 | 0.000215 | 0.000194 | 0.000200 |
