You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001418_02067
You are here: Home > Sequence: MGYG000001418_02067
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
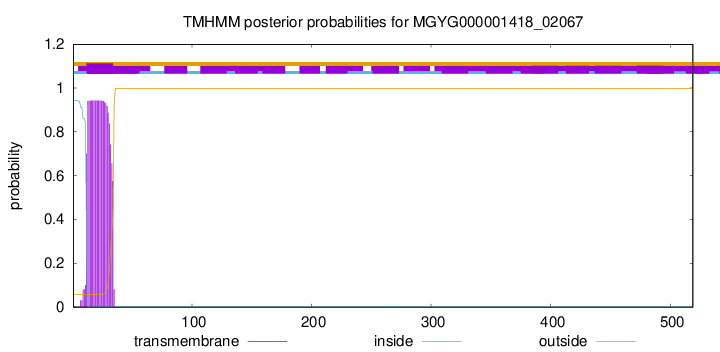
TMHMM annotations
Basic Information help
| Species | Aeromicrobium massiliense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Propionibacteriales; Nocardioidaceae; Aeromicrobium; Aeromicrobium massiliense | |||||||||||
| CAZyme ID | MGYG000001418_02067 | |||||||||||
| CAZy Family | CBM5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 641031; End: 642590 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| sd00042 | LVIVD | 2.14e-14 | 109 | 211 | 1 | 84 | LVIVD repeat. LVIVD repeats are mainly found in bacterial and archaeal cell surface proteins, many of them hypothetical. Structurally, LVIVD repeats have been predicted to form a beta-propeller, with each repeat forming one four-stranded anti-parallel beta-sheet blade. |
| sd00042 | LVIVD | 1.30e-11 | 98 | 205 | 33 | 120 | LVIVD repeat. LVIVD repeats are mainly found in bacterial and archaeal cell surface proteins, many of them hypothetical. Structurally, LVIVD repeats have been predicted to form a beta-propeller, with each repeat forming one four-stranded anti-parallel beta-sheet blade. |
| COG5276 | COG5276 | 4.62e-06 | 66 | 212 | 80 | 218 | Uncharacterized conserved protein [Function unknown]. |
| COG5276 | COG5276 | 5.07e-04 | 98 | 199 | 165 | 267 | Uncharacterized conserved protein [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARU51130.1 | 2.85e-209 | 74 | 519 | 54 | 484 |
| QDP73908.1 | 1.22e-208 | 15 | 519 | 21 | 505 |
| ARK04236.1 | 4.94e-208 | 15 | 519 | 21 | 505 |
| QDW61713.1 | 9.32e-204 | 49 | 517 | 44 | 496 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
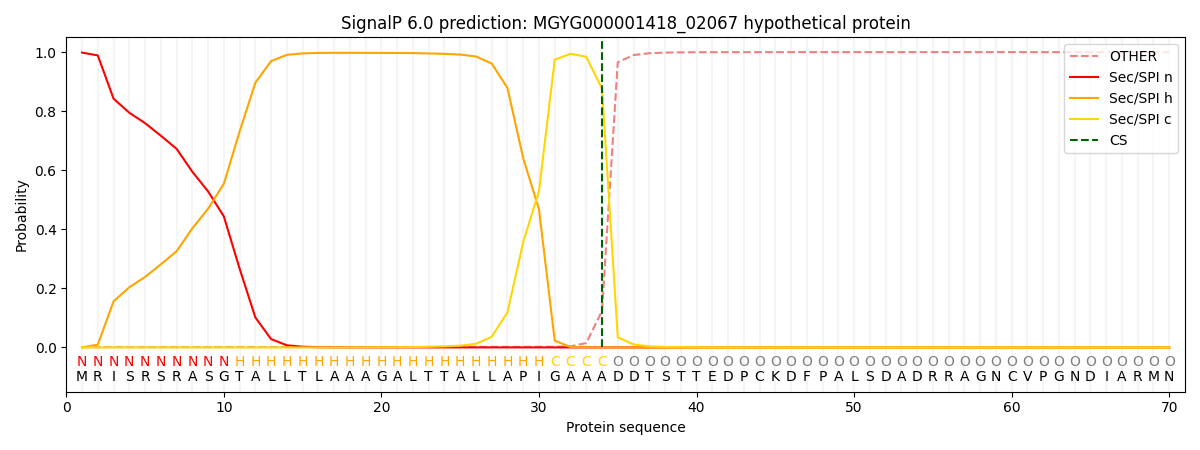
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001898 | 0.996152 | 0.000235 | 0.001228 | 0.000240 | 0.000194 |