You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001420_00174
You are here: Home > Sequence: MGYG000001420_00174
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes senegalensis | |||||||||||
| CAZyme ID | MGYG000001420_00174 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 221716; End: 224724 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.27e-22 | 56 | 481 | 59 | 494 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.42e-15 | 29 | 349 | 15 | 352 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 9.62e-14 | 21 | 352 | 87 | 397 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam02836 | Glyco_hydro_2_C | 2.31e-05 | 270 | 391 | 2 | 135 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam00703 | Glyco_hydro_2 | 1.01e-04 | 179 | 267 | 14 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY51369.1 | 0.0 | 20 | 983 | 22 | 994 |
| QNN44806.1 | 0.0 | 26 | 978 | 33 | 992 |
| QEC78585.1 | 0.0 | 27 | 983 | 33 | 996 |
| BCI63942.1 | 0.0 | 7 | 982 | 7 | 981 |
| QJC53152.1 | 9.56e-22 | 25 | 468 | 19 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1BHG_A | 5.24e-10 | 33 | 408 | 49 | 447 | HumanBeta-Glucuronidase At 2.6 A Resolution [Homo sapiens],1BHG_B Human Beta-Glucuronidase At 2.6 A Resolution [Homo sapiens],3HN3_A Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_B Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_D Human beta-glucuronidase at 1.7 A resolution [Homo sapiens],3HN3_E Human beta-glucuronidase at 1.7 A resolution [Homo sapiens] |
| 6NCW_A | 2.34e-08 | 60 | 413 | 54 | 402 | Crystalstructure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_B Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_C Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_D Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCX_A Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_B Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_C Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_D Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi] |
| 5UJ6_A | 2.91e-08 | 60 | 408 | 76 | 420 | CrystalStructure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],5UJ6_B Crystal Structure of Bacteroides Uniformis beta-glucuronidase [Bacteroides uniformis str. 3978 T3 ii],6NZG_A Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis],6NZG_B Bacteroides uniformis beta-glucuronidase 2 covalently bound to cyclophellitol-6-carboxylate aziridine [Bacteroides uniformis] |
| 6D50_A | 2.92e-08 | 60 | 408 | 84 | 428 | Bacteroidesuniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii],6D50_B Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone [Bacteroides uniformis str. 3978 T3 ii] |
| 3IAQ_A | 1.19e-07 | 138 | 392 | 192 | 462 | ChainA, Beta-galactosidase [Escherichia coli K-12],3IAQ_B Chain B, Beta-galactosidase [Escherichia coli K-12],3IAQ_C Chain C, Beta-galactosidase [Escherichia coli K-12],3IAQ_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q4FAT7 | 7.90e-13 | 60 | 401 | 98 | 461 | Beta-glucuronidase OS=Sus scrofa OX=9823 GN=GUSB PE=3 SV=1 |
| P06760 | 1.79e-12 | 42 | 413 | 71 | 454 | Beta-glucuronidase OS=Rattus norvegicus OX=10116 GN=Gusb PE=1 SV=1 |
| Q47077 | 4.15e-12 | 91 | 392 | 154 | 464 | Beta-galactosidase OS=Enterobacter cloacae OX=550 GN=lacZ PE=3 SV=1 |
| Q48846 | 9.05e-12 | 64 | 502 | 133 | 548 | Beta-galactosidase large subunit OS=Latilactobacillus sakei OX=1599 GN=lacL PE=3 SV=1 |
| A4W7D2 | 1.23e-11 | 91 | 392 | 154 | 464 | Beta-galactosidase OS=Enterobacter sp. (strain 638) OX=399742 GN=lacZ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
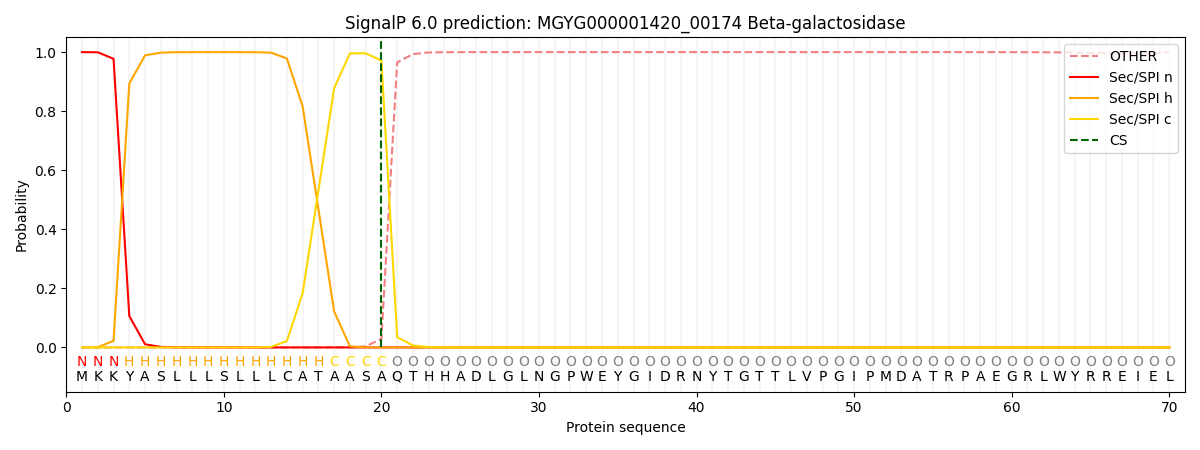
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006159 | 0.989512 | 0.001304 | 0.001108 | 0.000890 | 0.000970 |
