You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001420_00178
You are here: Home > Sequence: MGYG000001420_00178
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes senegalensis | |||||||||||
| CAZyme ID | MGYG000001420_00178 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 231382; End: 233970 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 19 | 462 | 2.9e-64 | 0.4933510638297872 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 9.18e-29 | 27 | 489 | 14 | 466 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 2.41e-22 | 114 | 462 | 141 | 481 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 5.58e-09 | 116 | 433 | 155 | 462 | beta-galactosidase. |
| pfam02836 | Glyco_hydro_2_C | 3.82e-06 | 310 | 453 | 1 | 157 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY53936.1 | 2.86e-282 | 20 | 854 | 451 | 1270 |
| AAU82837.1 | 1.82e-51 | 114 | 516 | 6 | 409 |
| QSR22851.1 | 7.64e-41 | 22 | 461 | 100 | 534 |
| QTD57081.1 | 9.75e-40 | 21 | 648 | 76 | 739 |
| QIJ31391.1 | 1.02e-35 | 2 | 585 | 7 | 612 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1YQ2_A | 7.64e-15 | 26 | 434 | 34 | 444 | ChainA, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_B Chain B, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_C Chain C, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_D Chain D, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_E Chain E, beta-galactosidase [Arthrobacter sp. C2-2],1YQ2_F Chain F, beta-galactosidase [Arthrobacter sp. C2-2] |
| 6ZJV_A | 1.72e-14 | 99 | 468 | 142 | 479 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A [Arthrobacter sp. 32cB],6ZJW_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with galactose [Arthrobacter sp. 32cB],6ZJX_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant D207A in complex with saccharose [Arthrobacter sp. 32cB] |
| 6ETZ_A | 2.24e-14 | 99 | 468 | 121 | 458 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB [Arthrobacter sp. 32cB],6H1P_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB - data collected at room temperature [Arthrobacter sp. 32cB] |
| 6SEB_A | 2.26e-14 | 99 | 468 | 142 | 479 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG [Arthrobacter sp. 32cB],6SEC_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG [Arthrobacter sp. 32cB],6SED_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose [Arthrobacter sp. 32cB] |
| 6ZJP_A | 2.26e-14 | 99 | 468 | 142 | 479 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q [Arthrobacter sp. 32cB] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q59140 | 6.89e-16 | 25 | 434 | 29 | 435 | Beta-galactosidase OS=Arthrobacter sp. (strain B7) OX=86041 GN=lacZ PE=1 SV=1 |
| B4S2K9 | 1.12e-12 | 117 | 579 | 155 | 616 | Beta-galactosidase OS=Alteromonas mediterranea (strain DSM 17117 / CIP 110805 / LMG 28347 / Deep ecotype) OX=1774373 GN=lacZ PE=3 SV=1 |
| Q8X685 | 5.06e-11 | 117 | 537 | 156 | 571 | Beta-galactosidase OS=Escherichia coli O157:H7 OX=83334 GN=lacZ PE=3 SV=1 |
| B5Z2P7 | 5.06e-11 | 117 | 537 | 156 | 571 | Beta-galactosidase OS=Escherichia coli O157:H7 (strain EC4115 / EHEC) OX=444450 GN=lacZ PE=3 SV=1 |
| A6TI29 | 5.06e-11 | 117 | 572 | 156 | 596 | Beta-galactosidase 2 OS=Klebsiella pneumoniae subsp. pneumoniae (strain ATCC 700721 / MGH 78578) OX=272620 GN=lacZ2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
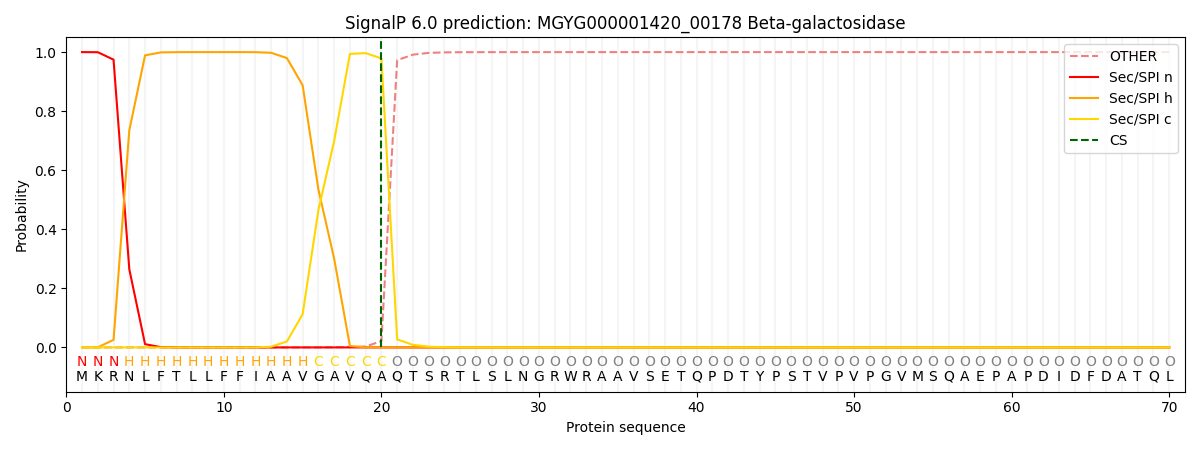
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000293 | 0.998946 | 0.000209 | 0.000186 | 0.000178 | 0.000158 |
