You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001420_00839
You are here: Home > Sequence: MGYG000001420_00839
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes senegalensis | |||||||||||
| CAZyme ID | MGYG000001420_00839 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1095005; End: 1096825 Strand: - | |||||||||||
Full Sequence Download help
| MRKTILTTVA ALSVLCTFAQ WKPAGDKLKT PWAEKIDPAN VLPEYPRPQL VRSQWMNLNG | 60 |
| LWDYAIKPAG ALEPKTMDGK ILVPFAVESS LSGVQKSLTE KDELWYRRTF SVPAAWKGSN | 120 |
| VMLNFGAVDW KADVFVNDIL VGSHTGGFTP FSLDITPCLK AKGDQKLVVR VFDGTDKGYQ | 180 |
| PRGKQALNPN GIWYTPVSGI WQTVWIEPVA KSHISGIKAI PNLDQKNIAV TVTADNCTTG | 240 |
| DLVEVNILDK GKVVASATGF PGSPLRLAIA EPKLWSPDSP FLYDMTAELK RGGKTVDKVD | 300 |
| SYTAMRKIGT ARDKAGILRM TLNDEPLFHY GPLDQGWWPD GLFTAPTDEA LLFDIVKAKE | 360 |
| LGYNMIRKHV KVEPARWYYH CDREGILVWQ DMPSGDMGNK WEMYIYNGGT DKLRTQESKD | 420 |
| NFSKEWKEIM DFCISSPSVV VWVPFNEAWG QFDTERIVNW TMEYDPSRLV NPASGGNHRP | 480 |
| CGHMLDLHHY PGPAMKLFDP QRVNVLGEYG GIGLALEGHL WWNKRNWGYV QFKNAEQVTA | 540 |
| EYEKYARNLA KFVRLGFSGA VYTQTTDVES EVNGLFTYDR KVMKVIPERI KAANEAVIRS | 600 |
| MSLEME | 606 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 50 | 483 | 3.9e-101 | 0.4920212765957447 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10150 | PRK10150 | 5.68e-34 | 47 | 471 | 3 | 449 | beta-D-glucuronidase; Provisional |
| COG3250 | LacZ | 1.54e-29 | 103 | 471 | 64 | 430 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02837 | Glyco_hydro_2_N | 8.45e-15 | 58 | 204 | 4 | 163 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 9.67e-15 | 106 | 308 | 113 | 320 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 1.11e-14 | 106 | 388 | 124 | 410 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK64944.1 | 0.0 | 1 | 606 | 1 | 606 |
| AST54135.1 | 0.0 | 1 | 602 | 1 | 603 |
| QUT94725.1 | 0.0 | 1 | 602 | 1 | 603 |
| BBK91716.1 | 0.0 | 1 | 602 | 1 | 603 |
| QIX63623.1 | 0.0 | 1 | 602 | 1 | 603 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7SF2_A | 1.51e-271 | 19 | 601 | 3 | 586 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_E Chain E, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838],7SF2_F Chain F, Glycosyl hydrolase family 2, sugar binding domain protein [Bacteroides cellulosilyticus DSM 14838] |
| 6ECA_A | 1.83e-24 | 58 | 472 | 43 | 463 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 4JKM_A | 1.24e-23 | 54 | 468 | 14 | 441 | CrystalStructure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],4JKM_B Crystal Structure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],6CXS_A Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13],6CXS_B Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13] |
| 6ED2_A | 7.49e-23 | 45 | 447 | 30 | 441 | ChainA, Glycosyl hydrolase family 2, TIM barrel domain protein [Faecalibacterium duncaniae] |
| 3GM8_A | 1.81e-22 | 105 | 470 | 69 | 421 | ChainA, Glycoside hydrolase family 2, candidate beta-glycosidase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P77989 | 4.09e-23 | 105 | 520 | 59 | 441 | Beta-galactosidase OS=Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E) OX=340099 GN=lacZ PE=3 SV=2 |
| T2KM09 | 3.93e-22 | 105 | 475 | 109 | 459 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| T2KPJ7 | 3.13e-21 | 105 | 456 | 106 | 439 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21970 PE=2 SV=1 |
| T2KN75 | 1.68e-18 | 104 | 466 | 89 | 457 | Beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22060 PE=1 SV=1 |
| P05804 | 3.40e-14 | 104 | 470 | 67 | 441 | Beta-glucuronidase OS=Escherichia coli (strain K12) OX=83333 GN=uidA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
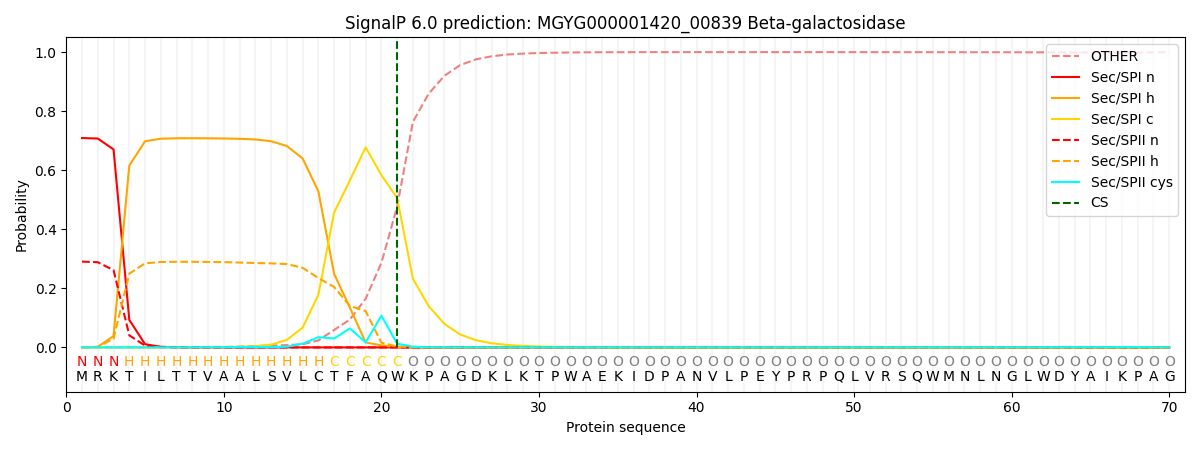
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001189 | 0.699902 | 0.298157 | 0.000312 | 0.000229 | 0.000206 |
