You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001420_03026
You are here: Home > Sequence: MGYG000001420_03026
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes senegalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes senegalensis | |||||||||||
| CAZyme ID | MGYG000001420_03026 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1581481; End: 1583214 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 227 | 529 | 6.9e-67 | 0.9839743589743589 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.73e-33 | 254 | 528 | 34 | 266 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.38e-10 | 226 | 435 | 53 | 274 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam16410 | DUF5018 | 0.002 | 28 | 205 | 42 | 215 | Domain of unknown function (DUF5018). This family of proteins is functionally uncharacterized. This family is found in various Bacteroides and Alistipes species. Proteins in this family are around 600 amino acids in length. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK63843.1 | 0.0 | 1 | 577 | 12 | 588 |
| AFL77440.1 | 0.0 | 1 | 577 | 1 | 577 |
| QMU29539.1 | 1.05e-58 | 220 | 560 | 40 | 385 |
| QNF33991.1 | 1.35e-55 | 222 | 561 | 45 | 389 |
| ACA61171.1 | 9.02e-53 | 216 | 560 | 31 | 384 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CEC_A | 1.15e-28 | 224 | 546 | 4 | 324 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 1CEN_A | 2.91e-28 | 224 | 546 | 4 | 324 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
| 6KDD_A | 1.63e-17 | 226 | 539 | 22 | 303 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 3W0K_A | 1.90e-16 | 255 | 546 | 33 | 313 | CrystalStructure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus],3W0K_B Crystal Structure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus] |
| 6XSU_A | 2.71e-09 | 246 | 529 | 52 | 314 | GH5-4broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens],6XSU_B GH5-4 broad specificity endoglucanase from Ruminococcus flavefaciens [Ruminococcus flavefaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DJ77 | 1.35e-28 | 224 | 546 | 4 | 324 | Endoglucanase C OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celC PE=3 SV=1 |
| P23340 | 1.35e-28 | 224 | 546 | 4 | 324 | Endoglucanase C307 OS=Clostridium sp. (strain F1) OX=1508 GN=celC307 PE=1 SV=1 |
| P0C2S3 | 6.32e-28 | 224 | 546 | 4 | 324 | Endoglucanase C OS=Acetivibrio thermocellus OX=1515 GN=celC PE=1 SV=1 |
| P25472 | 1.07e-16 | 237 | 546 | 49 | 313 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
| Q8NKF9 | 1.92e-08 | 234 | 421 | 86 | 281 | Glucan 1,3-beta-glucosidase OS=Candida oleophila OX=45573 GN=EXG1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
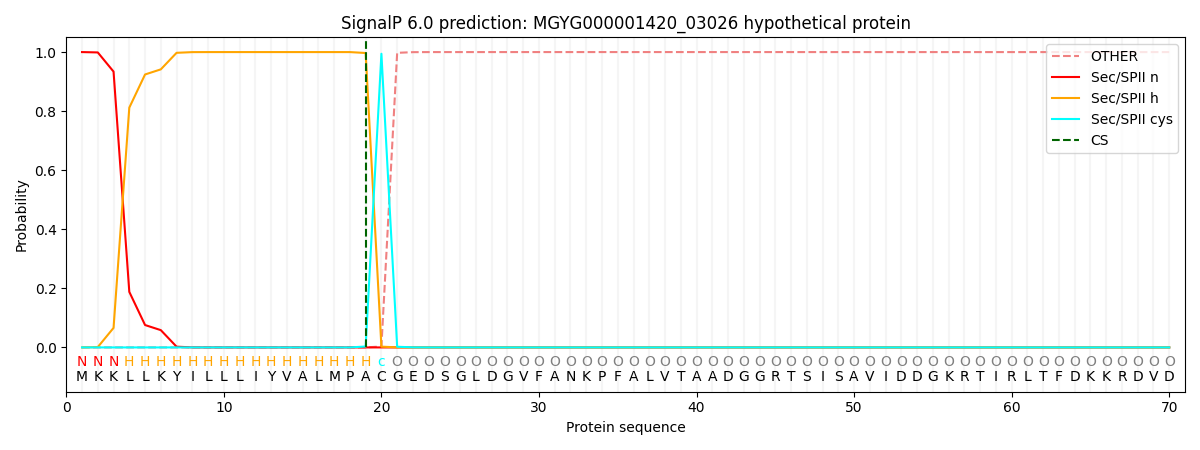
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000070 | 0.000000 | 0.000000 | 0.000000 |
