You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_02234
You are here: Home > Sequence: MGYG000001422_02234
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
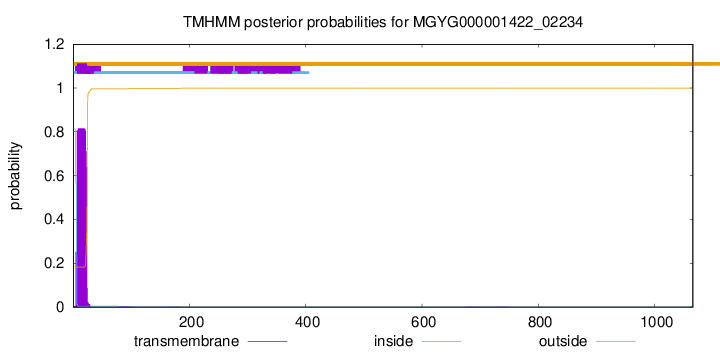
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_02234 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 718656; End: 721856 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH42 | 418 | 781 | 1.8e-84 | 0.9946091644204852 |
| GH105 | 73 | 404 | 3.9e-75 | 0.9728915662650602 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 4.85e-76 | 418 | 782 | 2 | 376 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| pfam07470 | Glyco_hydro_88 | 9.66e-75 | 48 | 405 | 1 | 342 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG1874 | GanA | 1.23e-61 | 407 | 1056 | 11 | 664 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| COG4225 | YesR | 5.02e-38 | 71 | 406 | 28 | 357 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| pfam08532 | Glyco_hydro_42M | 4.64e-22 | 811 | 1004 | 26 | 205 | Beta-galactosidase trimerisation domain. This is non catalytic domain B of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. This domain is related to glutamine amidotransferase enzymes, but the catalytic residues are replaced by non functional amino acids. This domain is involved in trimerisation. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEV98713.1 | 0.0 | 412 | 1066 | 22 | 677 |
| BAU54053.1 | 0.0 | 405 | 1065 | 10 | 671 |
| SDT03992.1 | 1.28e-315 | 405 | 1065 | 19 | 680 |
| AOW09973.1 | 9.02e-307 | 412 | 1065 | 25 | 676 |
| QUT76344.1 | 1.32e-188 | 40 | 427 | 31 | 413 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5NOA_A | 1.53e-60 | 44 | 409 | 32 | 378 | PolysaccharideLyase BACCELL_00875 [Bacteroides thetaiotaomicron] |
| 4Q88_A | 3.99e-60 | 72 | 407 | 32 | 359 | ChainA, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482],4Q88_B Chain B, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482] |
| 4UCF_A | 1.12e-57 | 412 | 1038 | 20 | 663 | Crystalstructure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_B Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UCF_C Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose [Bifidobacterium bifidum S17],4UZS_A Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_B Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17],4UZS_C Crystal structure of Bifidobacterium bifidum beta-galactosidase [Bifidobacterium bifidum S17] |
| 4UNI_A | 4.50e-56 | 410 | 904 | 21 | 532 | beta-(1,6)-galactosidasefrom Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UNI_B beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UNI_C beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04] |
| 4UOQ_A | 1.49e-55 | 410 | 904 | 21 | 532 | Nucleophilemutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis Bl-04],4UOQ_B Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis Bl-04],4UOQ_C Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis Bl-04],4UOZ_A beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UOZ_B beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04],4UOZ_C beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose [Bifidobacterium animalis subsp. lactis Bl-04] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D9SM34 | 3.85e-67 | 414 | 1064 | 3 | 657 | Beta-galactosidase BgaA OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=bgaA PE=1 SV=1 |
| P19668 | 3.21e-60 | 415 | 1025 | 10 | 623 | Beta-galactosidase bgaB OS=Geobacillus kaustophilus OX=1462 GN=bgaB PE=1 SV=1 |
| D5JGG0 | 3.85e-60 | 413 | 1064 | 9 | 662 | Beta-galactosidase LacZ OS=Weizmannia coagulans OX=1398 GN=lacZ PE=3 SV=1 |
| C9S0R2 | 1.99e-59 | 415 | 1025 | 10 | 623 | Beta-galactosidase BgaB OS=Geobacillus sp. (strain Y412MC61) OX=544556 GN=bgaB PE=3 SV=1 |
| D4QFE7 | 1.83e-57 | 412 | 1038 | 20 | 663 | Beta-galactosidase BbgII OS=Bifidobacterium bifidum OX=1681 GN=bbgII PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
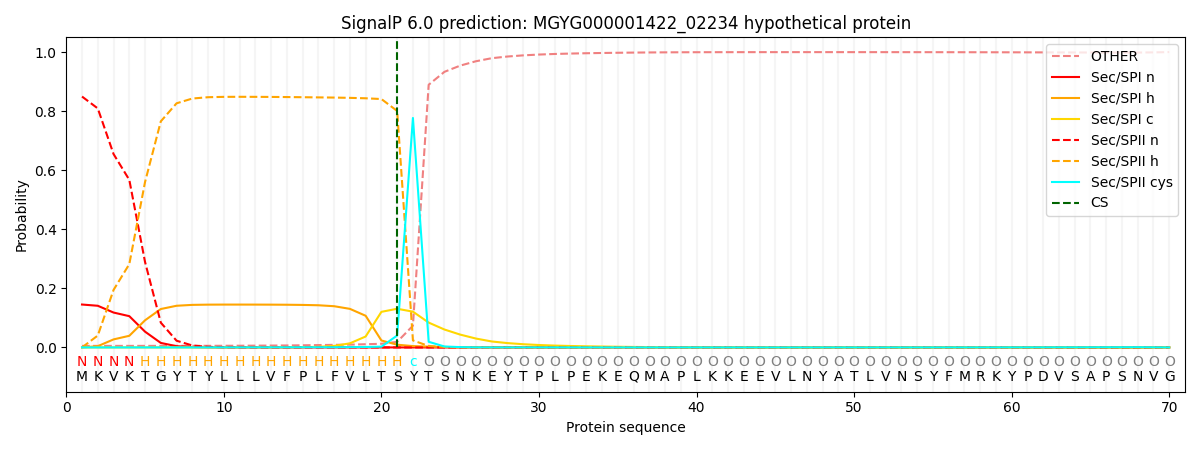
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.006862 | 0.139526 | 0.852969 | 0.000150 | 0.000214 | 0.000255 |