You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_02723
You are here: Home > Sequence: MGYG000001422_02723
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_02723 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1493708; End: 1496587 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 24 | 502 | 2.2e-36 | 0.5026595744680851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.58e-19 | 89 | 298 | 250 | 480 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 6.73e-12 | 120 | 297 | 1 | 181 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10340 | ebgA | 1.04e-11 | 78 | 272 | 272 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam16353 | DUF4981 | 0.004 | 461 | 514 | 11 | 64 | Domain of unknown function(DUF4981). This family consists of uncharacterized proteins around 1000 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
| pfam00703 | Glyco_hydro_2 | 0.005 | 24 | 118 | 7 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO68496.1 | 0.0 | 1 | 959 | 1 | 959 |
| QRQ49243.1 | 0.0 | 1 | 953 | 1 | 953 |
| QUT44989.1 | 0.0 | 1 | 953 | 1 | 953 |
| QDH57419.1 | 0.0 | 1 | 954 | 1 | 954 |
| AWO00586.1 | 1.63e-247 | 80 | 948 | 84 | 947 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 4.21e-13 | 77 | 290 | 269 | 482 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 4.21e-13 | 77 | 290 | 270 | 483 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 4JKM_A | 7.10e-11 | 91 | 297 | 245 | 475 | CrystalStructure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],4JKM_B Crystal Structure of Clostridium perfringens beta-glucuronidase [Clostridium perfringens str. 13],6CXS_A Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13],6CXS_B Crystal Structure of Clostridium perfringens beta-glucuronidase bound with a novel, potent inhibitor 4-(8-(piperazin-1-yl)-1,2,3,4-tetrahydro-[1,2,3]triazino[4',5':4,5]thieno[2,3-c]isoquinolin-5-yl)morpholine [Clostridium perfringens str. 13] |
| 6BO6_A | 1.20e-07 | 91 | 298 | 258 | 486 | Eubacteriumeligens beta-glucuronidase bound to UNC4917 glucuronic acid conjugate [[Eubacterium] eligens ATCC 27750] |
| 6D4O_A | 1.20e-07 | 91 | 298 | 261 | 489 | Eubacteriumeligens beta-glucuronidase bound to an amoxapine-glucuronide conjugate [Lachnospira eligens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.31e-12 | 77 | 290 | 270 | 483 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q03WL0 | 7.81e-11 | 77 | 254 | 282 | 468 | Beta-galactosidase OS=Leuconostoc mesenteroides subsp. mesenteroides (strain ATCC 8293 / DSM 20343 / BCRC 11652 / CCM 1803 / JCM 6124 / NCDO 523 / NBRC 100496 / NCIMB 8023 / NCTC 12954 / NRRL B-1118 / 37Y) OX=203120 GN=lacZ PE=3 SV=1 |
| Q9K9C6 | 1.79e-08 | 74 | 272 | 276 | 482 | Beta-galactosidase OS=Alkalihalobacillus halodurans (strain ATCC BAA-125 / DSM 18197 / FERM 7344 / JCM 9153 / C-125) OX=272558 GN=lacZ PE=3 SV=1 |
| Q6LL68 | 1.80e-08 | 57 | 253 | 261 | 467 | Beta-galactosidase OS=Photobacterium profundum (strain SS9) OX=298386 GN=lacZ PE=3 SV=1 |
| Q8D4H3 | 1.59e-07 | 77 | 253 | 283 | 469 | Beta-galactosidase OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=lacZ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
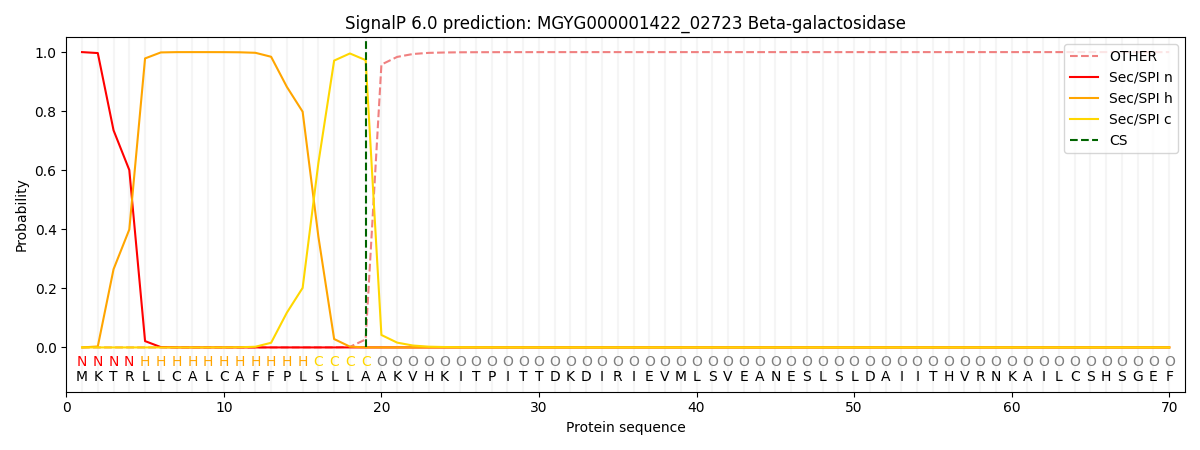
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000364 | 0.998955 | 0.000182 | 0.000167 | 0.000150 | 0.000146 |
