You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_03090
You are here: Home > Sequence: MGYG000001422_03090
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
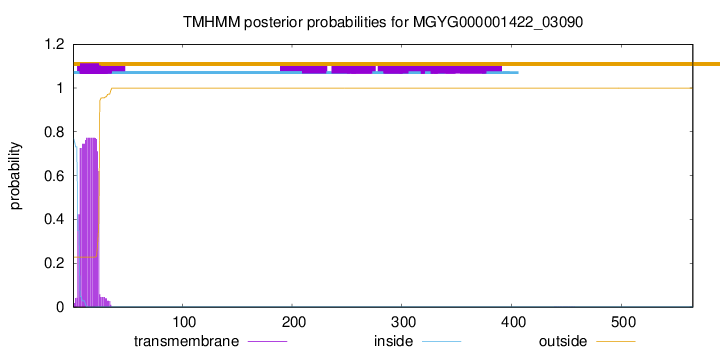
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_03090 | |||||||||||
| CAZy Family | CE12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 403863; End: 405560 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE12 | 30 | 251 | 4.4e-76 | 0.9904761904761905 |
| CE4 | 279 | 401 | 6.8e-28 | 0.8538461538461538 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01821 | Rhamnogalacturan_acetylesterase_like | 1.75e-86 | 29 | 252 | 1 | 198 | Rhamnogalacturan_acetylesterase_like subgroup of SGNH-hydrolases. Rhamnogalacturan acetylesterase removes acetyl esters from rhamnogalacturonan substrates, and renders them susceptible to degradation by rhamnogalacturonases. Rhamnogalacturonans are highly branched regions in pectic polysaccharides, consisting of repeating -(1,2)-L-Rha-(1,4)-D-GalUA disaccharide units, with many rhamnose residues substituted by neutral oligosaccharides such as arabinans, galactans and arabinogalactans. Extracellular enzymes participating in the degradation of plant cell wall polymers, such as Rhamnogalacturonan acetylesterase, would typically be found in saprophytic and plant pathogenic fungi and bacteria. |
| cd10918 | CE4_NodB_like_5s_6s | 5.21e-32 | 286 | 399 | 5 | 123 | Putative catalytic NodB homology domain of PgaB, IcaB, and similar proteins which consist of a deformed (beta/alpha)8 barrel fold with 5- or 6-strands. This family belongs to the large and functionally diverse carbohydrate esterase 4 (CE4) superfamily, whose members show strong sequence similarity with some variability due to their distinct carbohydrate substrates. It includes bacterial poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase PgaB, hemin storage system HmsF protein in gram-negative species, intercellular adhesion proteins IcaB, and many uncharacterized prokaryotic polysaccharide deacetylases. It also includes a putative polysaccharide deacetylase YxkH encoded by the Bacillus subtilis yxkH gene, which is one of six polysaccharide deacetylase gene homologs present in the Bacillus subtilis genome. Sequence comparison shows all family members contain a conserved domain similar to the catalytic NodB homology domain of rhizobial NodB-like proteins, which consists of a deformed (beta/alpha)8 barrel fold with 6 or 7 strands. However, in this family, most proteins have 5 strands and some have 6 strands. Moreover, long insertions are found in many family members, whose function remains unknown. |
| cd10970 | CE4_DAC_u1_6s | 6.90e-32 | 282 | 458 | 2 | 186 | Putative catalytic NodB homology domain of uncharacterized prokaryotic polysaccharide deacetylases which consist of a 6-stranded beta/alpha barrel. This family contains uncharacterized prokaryotic polysaccharide deacetylases. Although their biological functions remain unknown, all members of the family contain a conserved domain with a 6-stranded beta/alpha barrel, which is similar to the catalytic NodB homology domain of rhizobial NodB-like proteins, belonging to the larger carbohydrate esterase 4 (CE4) superfamily. |
| cd10967 | CE4_GLA_like_6s | 4.49e-30 | 281 | 469 | 1 | 202 | Putative catalytic NodB homology domain of gellan lyase and similar proteins. This family is represented by the extracellular polysaccharide-degrading enzyme, gellan lyase (gellanase, EC 4.2.2.-), from Bacillus sp. The enzyme acts on gellan exolytically and releases a tetrasaccharide of glucuronyl-glucosyl-rhamnosyl-glucose with unsaturated glucuronic acid at the nonreducing terminus. The family also includes many uncharacterized prokaryotic polysaccharide deacetylases, which show high sequence similarity to Bacillus sp. gellan lyase. Although their biological functions remain unknown, all members of the family contain a conserved domain with a 6-stranded beta/alpha barrel, which is similar to the catalytic NodB homology domain of rhizobial NodB-like proteins, belonging to the larger carbohydrate esterase 4 (CE4) superfamily. |
| pfam01522 | Polysacc_deac_1 | 3.20e-25 | 286 | 402 | 12 | 118 | Polysaccharide deacetylase. This domain is found in polysaccharide deacetylase. This family of polysaccharide deacetylases includes NodB (nodulation protein B from Rhizobium) which is a chitooligosaccharide deacetylase. It also includes chitin deacetylase from yeast, and endoxylanases which hydrolyzes glucosidic bonds in xylan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO67467.1 | 0.0 | 1 | 565 | 1 | 562 |
| QQY38016.1 | 0.0 | 1 | 565 | 1 | 565 |
| ABR39821.1 | 0.0 | 1 | 565 | 1 | 565 |
| QUT68402.1 | 0.0 | 1 | 565 | 1 | 565 |
| QDM12561.1 | 0.0 | 4 | 565 | 1 | 562 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2O14_A | 2.86e-16 | 30 | 254 | 164 | 361 | X-RayCrystal Structure of Protein YXIM_BACsu from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR595 [Bacillus subtilis] |
| 1DEO_A | 6.13e-14 | 30 | 252 | 2 | 212 | RHAMNOGALACTURONANACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.55 A RESOLUTION WITH SO4 IN THE ACTIVE SITE [Aspergillus aculeatus],1DEX_A RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.9 A RESOLUTION [Aspergillus aculeatus],1K7C_A Rhamnogalacturonan acetylesterase with seven N-linked carbohydrate residues distributed at two N-glycosylation sites refined at 1.12 A resolution [Aspergillus aculeatus],1PP4_A The crystal structure of rhamnogalacturonan acetylesterase in space group P3121 [Aspergillus aculeatus],1PP4_B The crystal structure of rhamnogalacturonan acetylesterase in space group P3121 [Aspergillus aculeatus] |
| 3C1U_A | 2.77e-13 | 30 | 252 | 2 | 212 | ChainA, Rhamnogalacturonan acetylesterase [Aspergillus aculeatus] |
| 7FBW_A | 1.39e-10 | 283 | 426 | 119 | 252 | ChainA, Predicted xylanase/chitin deacetylase [Caldanaerobacter subterraneus subsp. tengcongensis MB4] |
| 7BLY_A | 8.55e-08 | 282 | 437 | 41 | 189 | ChainA, Aspergillus niger contig An12c0130, genomic contig [Aspergillus niger CBS 513.88] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31523 | 1.37e-40 | 26 | 252 | 3 | 212 | Rhamnogalacturonan acetylesterase RhgT OS=Bacillus subtilis (strain 168) OX=224308 GN=rhgT PE=1 SV=1 |
| O31528 | 2.46e-25 | 31 | 259 | 5 | 205 | Probable rhamnogalacturonan acetylesterase YesY OS=Bacillus subtilis (strain 168) OX=224308 GN=yesY PE=1 SV=1 |
| P42304 | 1.55e-17 | 30 | 254 | 179 | 376 | Uncharacterized esterase YxiM OS=Bacillus subtilis (strain 168) OX=224308 GN=yxiM PE=1 SV=2 |
| Q00017 | 4.41e-13 | 30 | 252 | 19 | 229 | Rhamnogalacturonan acetylesterase OS=Aspergillus aculeatus OX=5053 GN=rha1 PE=1 SV=1 |
| Q5BAA2 | 7.44e-13 | 30 | 252 | 19 | 224 | Rhamnogalacturonan acetylesterase OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=AN2528 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
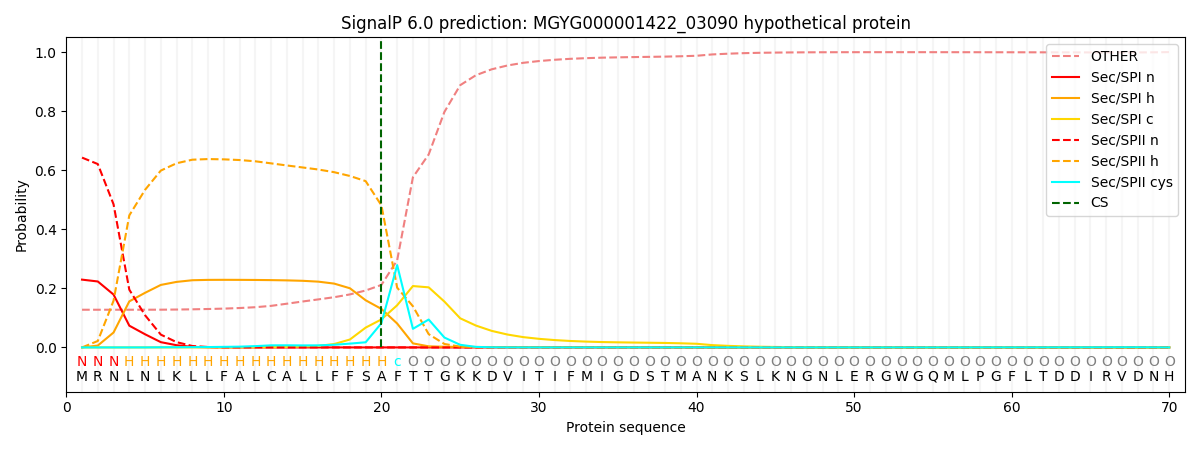
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.130703 | 0.222951 | 0.645653 | 0.000159 | 0.000166 | 0.000353 |