You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_03889
You are here: Home > Sequence: MGYG000001422_03889
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_03889 | |||||||||||
| CAZy Family | GH105 | |||||||||||
| CAZyme Description | Unsaturated rhamnogalacturonyl hydrolase YteR | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 632029; End: 633339 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 99 | 429 | 2.7e-75 | 0.9668674698795181 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 7.15e-63 | 84 | 430 | 6 | 341 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 2.94e-56 | 75 | 375 | 7 | 300 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| cd04791 | LanC_SerThrkinase | 9.71e-06 | 69 | 249 | 100 | 290 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
| COG1331 | YyaL | 0.002 | 101 | 194 | 469 | 582 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| COG1331 | YyaL | 0.002 | 100 | 193 | 407 | 511 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALJ58448.1 | 1.01e-291 | 1 | 434 | 1 | 434 |
| QUT90439.1 | 1.43e-291 | 1 | 434 | 1 | 434 |
| QDO71120.1 | 1.17e-290 | 1 | 434 | 1 | 434 |
| AWI10254.1 | 1.10e-127 | 56 | 429 | 20 | 406 |
| QMW80470.1 | 3.66e-80 | 80 | 429 | 7 | 358 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4WU0_A | 1.08e-50 | 104 | 430 | 25 | 360 | StructuralAnalysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824],4WU0_B Structural Analysis of C. acetobutylicum ATCC 824 Glycoside Hydrolase From Family 105 [Clostridium acetobutylicum ATCC 824] |
| 1NC5_A | 5.80e-49 | 75 | 395 | 12 | 332 | Structureof Protein of Unknown Function of YteR from Bacillus Subtilis [Bacillus subtilis],2D8L_A Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc [Bacillus subtilis] |
| 2GH4_A | 2.44e-48 | 75 | 395 | 2 | 322 | ChainA, Putative glycosyl hydrolase yteR [Bacillus subtilis] |
| 3K11_A | 7.47e-17 | 103 | 432 | 81 | 418 | Crystalstructure of Putative glycosyl hydrolase (NP_813087.1) from BACTEROIDES THETAIOTAOMICRON VPI-5482 at 1.80 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4Q88_A | 8.87e-06 | 119 | 375 | 54 | 307 | ChainA, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482],4Q88_B Chain B, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34559 | 3.18e-48 | 75 | 395 | 12 | 332 | Unsaturated rhamnogalacturonyl hydrolase YteR OS=Bacillus subtilis (strain 168) OX=224308 GN=yteR PE=1 SV=1 |
| P0A3U6 | 2.24e-13 | 213 | 391 | 5 | 183 | Protein Atu3128 OS=Agrobacterium fabrum (strain C58 / ATCC 33970) OX=176299 GN=Atu3128 PE=3 SV=1 |
| P0A3U7 | 2.24e-13 | 213 | 391 | 5 | 183 | 24.9 kDa protein in picA locus OS=Rhizobium radiobacter OX=358 PE=2 SV=1 |
| P9WF04 | 8.48e-09 | 265 | 375 | 257 | 365 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_28 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
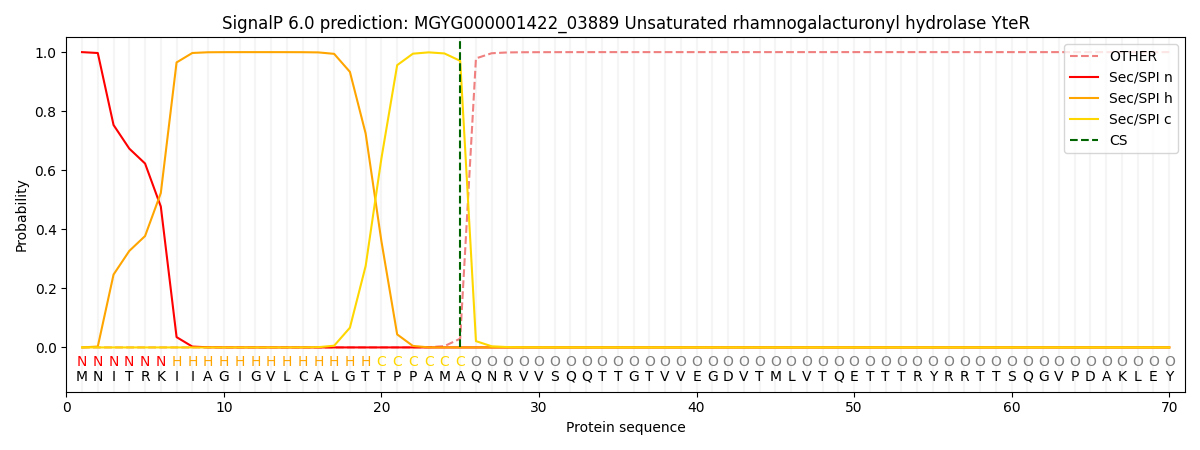
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000569 | 0.997770 | 0.000945 | 0.000269 | 0.000237 | 0.000197 |
