You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_04530
You are here: Home > Sequence: MGYG000001422_04530
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_04530 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 633532; End: 634926 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 23 | 421 | 2.1e-35 | 0.845213849287169 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 2.54e-07 | 104 | 241 | 1 | 116 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 2.37e-05 | 96 | 223 | 16 | 155 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 3.89e-05 | 133 | 320 | 1 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 8.24e-05 | 156 | 342 | 1 | 145 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT44652.1 | 0.0 | 1 | 464 | 1 | 464 |
| QUT88190.1 | 1.08e-274 | 1 | 461 | 1 | 460 |
| BBE17123.1 | 6.56e-127 | 1 | 390 | 1 | 397 |
| QEC62640.1 | 2.04e-100 | 8 | 415 | 17 | 422 |
| ASU33528.1 | 3.43e-92 | 23 | 419 | 33 | 427 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
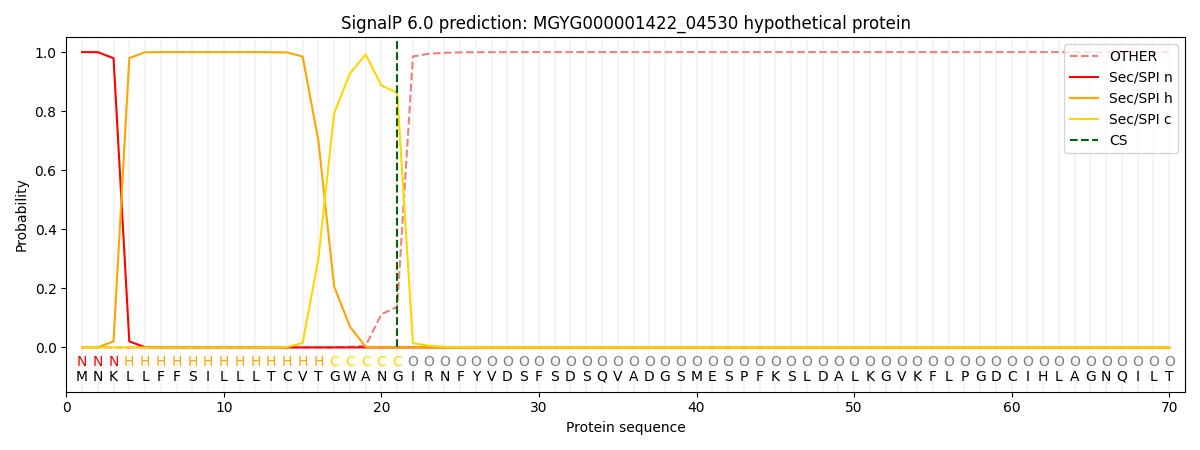
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000238 | 0.999174 | 0.000156 | 0.000145 | 0.000136 | 0.000128 |
