You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001422_05328
You are here: Home > Sequence: MGYG000001422_05328
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
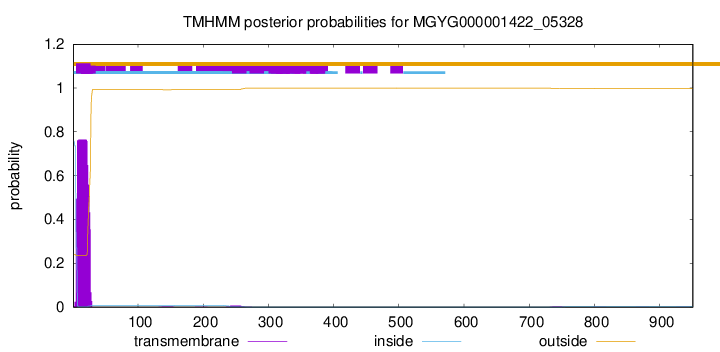
TMHMM annotations
Basic Information help
| Species | Bacteroides oleiciplenus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides oleiciplenus | |||||||||||
| CAZyme ID | MGYG000001422_05328 | |||||||||||
| CAZy Family | PL35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 352393; End: 355248 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 631 | 944 | 4.2e-88 | 0.9728915662650602 |
| PL35 | 385 | 560 | 1.6e-58 | 0.9664804469273743 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 7.05e-93 | 617 | 945 | 3 | 342 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| COG4225 | YesR | 7.19e-61 | 624 | 946 | 23 | 357 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| pfam07940 | Hepar_II_III | 2.07e-07 | 385 | 586 | 27 | 220 | Heparinase II/III-like protein. This family features sequences that are similar to a region of the Flavobacterium heparinum proteins heparinase II and heparinase III. The former is known to degrade heparin and heparin sulphate, whereas the latter predominantly degrades heparin sulphate. Both are secreted into the periplasmic space upon induction with heparin. |
| pfam16332 | DUF4962 | 8.58e-04 | 33 | 322 | 136 | 444 | Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ48160.1 | 0.0 | 7 | 945 | 7 | 946 |
| QUT46248.1 | 0.0 | 86 | 945 | 1 | 860 |
| AUD03829.1 | 3.27e-182 | 7 | 622 | 10 | 626 |
| QHV93651.1 | 6.59e-181 | 1 | 611 | 1 | 614 |
| ADB38475.1 | 2.07e-179 | 21 | 608 | 28 | 611 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CE7_A | 6.17e-75 | 633 | 945 | 49 | 369 | Crystalstructure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans],4CE7_B Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans],4CE7_C Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation [Nonlabens ulvanivorans] |
| 5NOA_A | 4.98e-56 | 622 | 941 | 44 | 370 | PolysaccharideLyase BACCELL_00875 [Bacteroides thetaiotaomicron] |
| 4Q88_A | 4.32e-43 | 633 | 941 | 63 | 353 | ChainA, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482],4Q88_B Chain B, Uncharacterized protein [Phocaeicola vulgatus ATCC 8482] |
| 1NC5_A | 1.50e-15 | 640 | 951 | 44 | 373 | Structureof Protein of Unknown Function of YteR from Bacillus Subtilis [Bacillus subtilis],2D8L_A Crystal Structure of Unsaturated Rhamnogalacturonyl Hydrolase in complex with dGlcA-GalNAc [Bacillus subtilis] |
| 2GH4_A | 5.84e-15 | 640 | 951 | 34 | 363 | ChainA, Putative glycosyl hydrolase yteR [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KPL9 | 1.13e-74 | 636 | 947 | 57 | 377 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22220 PE=2 SV=1 |
| L7P9J4 | 4.16e-74 | 633 | 945 | 56 | 376 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Nonlabens ulvanivorans OX=906888 GN=IL45_01505 PE=1 SV=1 |
| P9WF04 | 2.81e-70 | 613 | 943 | 58 | 409 | Unsaturated 3S-rhamnoglycuronyl hydrolase OS=Alteromonas sp. (strain LOR) OX=1537994 GN=LOR_28 PE=1 SV=1 |
| O34559 | 8.23e-15 | 640 | 951 | 44 | 373 | Unsaturated rhamnogalacturonyl hydrolase YteR OS=Bacillus subtilis (strain 168) OX=224308 GN=yteR PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
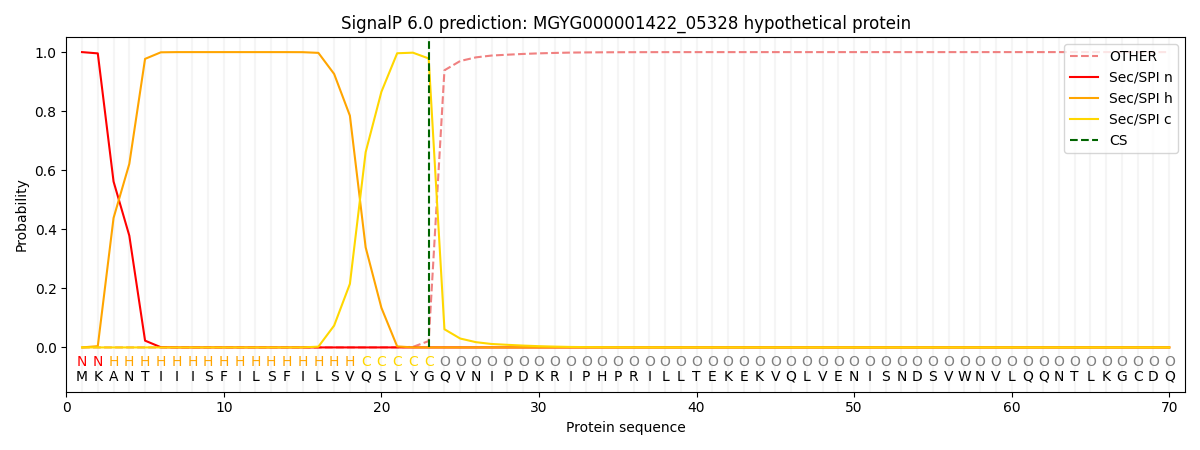
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000224 | 0.999203 | 0.000130 | 0.000153 | 0.000131 | 0.000128 |