You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001423_03227
You are here: Home > Sequence: MGYG000001423_03227
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
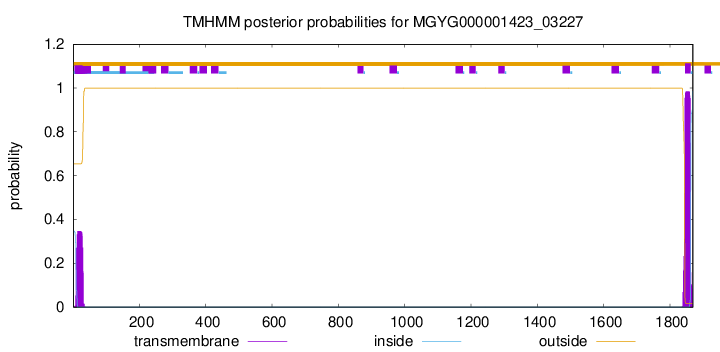
TMHMM annotations
Basic Information help
| Species | Clostridium celatum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium; Clostridium celatum | |||||||||||
| CAZyme ID | MGYG000001423_03227 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 15077; End: 20686 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 108 | 246 | 4e-23 | 0.967741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.92e-23 | 107 | 246 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.56e-10 | 106 | 233 | 12 | 130 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 1.80e-08 | 1187 | 1304 | 7 | 120 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02368 | Big_2 | 1.32e-06 | 1495 | 1562 | 2 | 70 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG0419 | SbcC | 2.17e-06 | 1566 | 1773 | 466 | 663 | DNA repair exonuclease SbcCD ATPase subunit [Replication, recombination and repair]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCC62414.1 | 2.91e-288 | 77 | 1511 | 3 | 1434 |
| SQG39183.1 | 5.63e-237 | 30 | 1326 | 35 | 1367 |
| ALG48827.1 | 2.99e-236 | 1 | 1326 | 6 | 1367 |
| AXH52486.1 | 5.82e-236 | 1 | 1326 | 6 | 1367 |
| BAB80970.1 | 4.30e-235 | 1 | 1326 | 6 | 1367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2J7M_A | 1.44e-11 | 109 | 249 | 14 | 148 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 1.48e-11 | 109 | 249 | 15 | 149 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
| 1K3I_A | 6.88e-10 | 78 | 248 | 3 | 162 | CrystalStructure of the Precursor of Galactose Oxidase [Fusarium sp.] |
| 2V5D_A | 7.41e-10 | 109 | 249 | 602 | 736 | Structureof a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. [Clostridium perfringens] |
| 2EIC_A | 4.57e-09 | 93 | 248 | 1 | 145 | ChainA, Galactose oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I1S2N3 | 5.72e-10 | 78 | 248 | 27 | 186 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0DTR4 | 1.07e-09 | 101 | 249 | 509 | 644 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q0TR53 | 1.58e-09 | 109 | 259 | 632 | 776 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 2.07e-09 | 109 | 278 | 632 | 795 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| P0CS93 | 3.86e-09 | 78 | 248 | 27 | 186 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
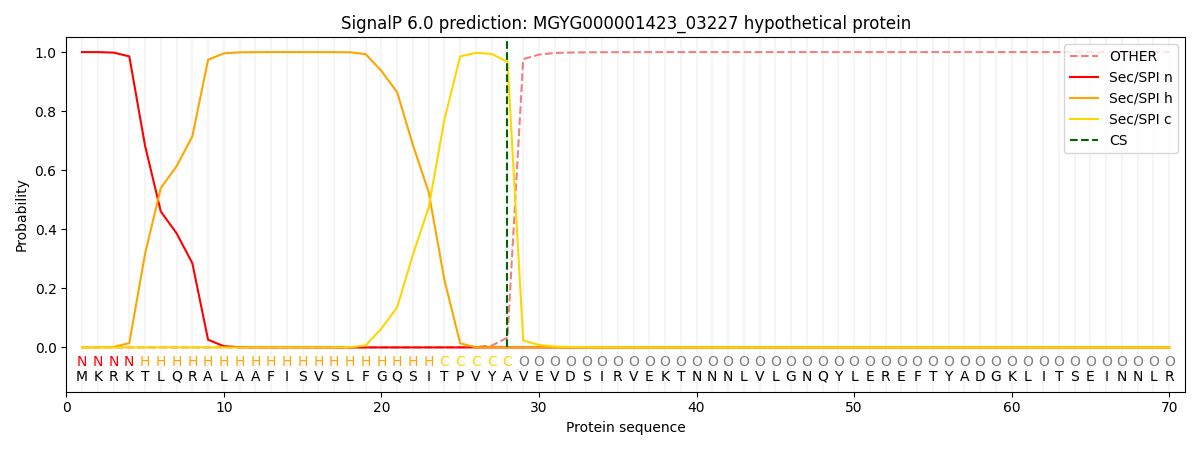
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000306 | 0.998936 | 0.000239 | 0.000186 | 0.000180 | 0.000159 |