You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001426_00810
You are here: Home > Sequence: MGYG000001426_00810
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytobacter massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Phytobacter; Phytobacter massiliensis | |||||||||||
| CAZyme ID | MGYG000001426_00810 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | Periplasmic trehalase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 852162; End: 853847 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH37 | 58 | 533 | 3.3e-187 | 0.9959266802443992 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13270 | treF | 0.0 | 49 | 533 | 62 | 545 | alpha,alpha-trehalase TreF. |
| PRK13272 | treA | 0.0 | 19 | 537 | 22 | 539 | alpha,alpha-trehalase TreA. |
| COG1626 | TreA | 0.0 | 36 | 540 | 48 | 558 | Neutral trehalase [Carbohydrate transport and metabolism]. |
| PRK13271 | treA | 0.0 | 1 | 561 | 1 | 565 | alpha,alpha-trehalase TreA. |
| pfam01204 | Trehalase | 0.0 | 57 | 533 | 1 | 507 | Trehalase. Trehalase (EC:3.2.1.28) is known to recycle trehalose to glucose. Trehalose is a physiological hallmark of heat-shock response in yeast and protects of proteins and membranes against a variety of stresses. This family is found in conjunction with pfam07492 in fungi. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJF18262.1 | 0.0 | 1 | 561 | 1 | 564 |
| QOV70633.1 | 0.0 | 1 | 561 | 1 | 564 |
| AUV09415.1 | 0.0 | 1 | 561 | 1 | 564 |
| AUU90498.1 | 0.0 | 1 | 561 | 1 | 564 |
| AUV01445.1 | 0.0 | 1 | 561 | 1 | 564 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JG0_A | 0.0 | 34 | 554 | 2 | 522 | Family37 trehalase from Escherichia coli in complex with 1- thiatrehazolin [Escherichia coli K-12],2JJB_A Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_B Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_C Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2JJB_D Family 37 trehalase from Escherichia coli in complex with casuarine-6- O-alpha-glucopyranose [Escherichia coli K-12],2WYN_A Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_B Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_C Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12],2WYN_D Structure of family 37 trehalase from Escherichia coli in complex with a casuarine-6-O-a-D-glucoside analogue [Escherichia coli K-12] |
| 5Z66_A | 0.0 | 41 | 551 | 46 | 556 | Structureof periplasmic trehalase from Diamondback moth gut bacteria complexed with validoxylamine [Enterobacter cloacae],5Z6H_A Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form [Enterobacter cloacae],5Z6H_B Structure of periplasmic trehalase from Diamondback moth gut bacteria in the apo form [Enterobacter cloacae] |
| 2JF4_A | 1.35e-313 | 34 | 554 | 2 | 522 | Family37 trehalase from Escherichia coli in complex with validoxylamine [Escherichia coli K-12] |
| 7E9U_A | 1.97e-80 | 57 | 532 | 36 | 552 | ChainA, Trehalase [Arabidopsis thaliana],7E9U_B Chain B, Trehalase [Arabidopsis thaliana] |
| 7E9X_A | 2.91e-79 | 57 | 532 | 36 | 552 | ChainA, Trehalase [Arabidopsis thaliana],7E9X_B Chain B, Trehalase [Arabidopsis thaliana],7E9X_C Chain C, Trehalase [Arabidopsis thaliana],7E9X_D Chain D, Trehalase [Arabidopsis thaliana],7EAW_A Chain A, Trehalase [Arabidopsis thaliana],7EAW_B Chain B, Trehalase [Arabidopsis thaliana] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B7N408 | 0.0 | 30 | 556 | 24 | 554 | Periplasmic trehalase OS=Escherichia coli O17:K52:H18 (strain UMN026 / ExPEC) OX=585056 GN=treA PE=3 SV=1 |
| C4ZTN8 | 0.0 | 30 | 554 | 24 | 552 | Periplasmic trehalase OS=Escherichia coli (strain K12 / MC4100 / BW2952) OX=595496 GN=treA PE=3 SV=1 |
| C0Q337 | 0.0 | 4 | 554 | 4 | 559 | Periplasmic trehalase OS=Salmonella paratyphi C (strain RKS4594) OX=476213 GN=treA PE=3 SV=1 |
| B7NUW3 | 0.0 | 30 | 556 | 24 | 554 | Periplasmic trehalase OS=Escherichia coli O7:K1 (strain IAI39 / ExPEC) OX=585057 GN=treA PE=3 SV=1 |
| P13482 | 0.0 | 30 | 554 | 24 | 552 | Periplasmic trehalase OS=Escherichia coli (strain K12) OX=83333 GN=treA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
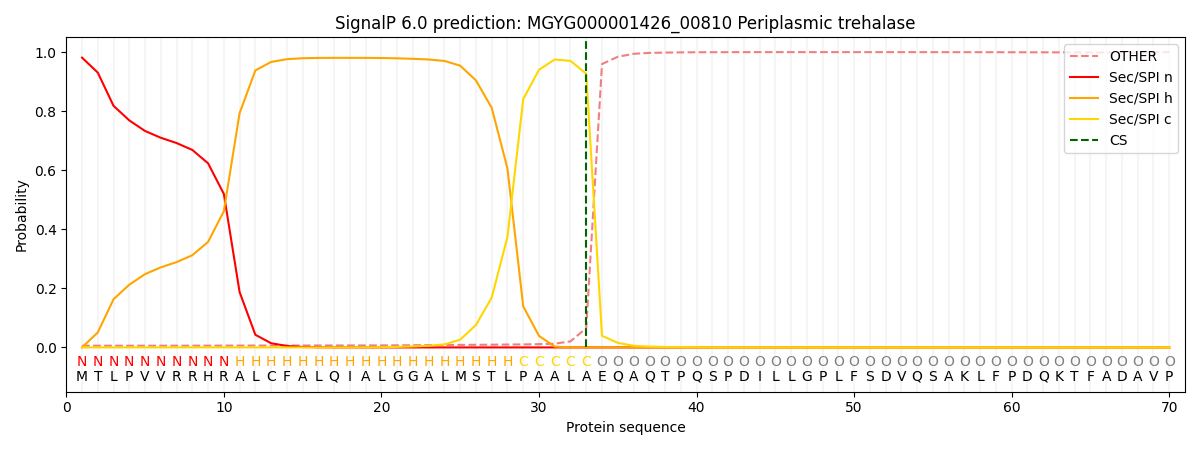
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.008799 | 0.975307 | 0.004836 | 0.010173 | 0.000530 | 0.000315 |
