You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001430_02356
You are here: Home > Sequence: MGYG000001430_02356
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
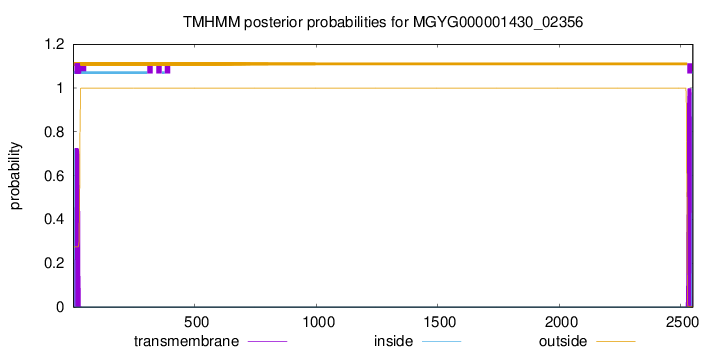
TMHMM annotations
Basic Information help
| Species | Holdemania massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Holdemania; Holdemania massiliensis | |||||||||||
| CAZyme ID | MGYG000001430_02356 | |||||||||||
| CAZy Family | PL8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 854312; End: 861964 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL8 | 1127 | 1408 | 9.5e-70 | 0.9884169884169884 |
| CBM32 | 1554 | 1692 | 1.2e-22 | 0.9354838709677419 |
| CBM32 | 1955 | 2084 | 2.7e-19 | 0.967741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01083 | GAG_Lyase | 8.42e-168 | 750 | 1479 | 1 | 693 | Glycosaminoglycan (GAG) polysaccharide lyase family. This family consists of a group of secreted bacterial lyase enzymes capable of acting on glycosaminoglycans, such as hyaluronan and chondroitin, in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. These are broad-specificity glycosaminoglycan lyases which recognize uronyl residues in polysaccharides and cleave their glycosidic bonds via a beta-elimination reaction to form a double bond between C-4 and C-5 of the non-reducing terminal uronyl residues of released products. Substrates include chondroitin, chondroitin 4-sulfate, chondroitin 6-sulfate, and hyaluronic acid. Family members include chondroitin AC lyase, chondroitin abc lyase, xanthan lyase, and hyalurate lyase. |
| pfam08124 | Lyase_8_N | 2.45e-74 | 758 | 1083 | 1 | 323 | Polysaccharide lyase family 8, N terminal alpha-helical domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| pfam02278 | Lyase_8 | 9.40e-66 | 1132 | 1407 | 7 | 252 | Polysaccharide lyase family 8, super-sandwich domain. This family consists of a group of secreted bacterial lyase enzymes EC:4.2.2.1 capable of acting on hyaluronan and chondroitin in the extracellular matrix of host tissues, contributing to the invasive capacity of the pathogen. |
| pfam02368 | Big_2 | 5.49e-18 | 1860 | 1936 | 1 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam00754 | F5_F8_type_C | 2.34e-15 | 1553 | 1691 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIZ07196.1 | 9.37e-111 | 740 | 1542 | 25 | 802 |
| AZS13398.1 | 3.10e-108 | 405 | 1540 | 32 | 1066 |
| BBH21832.1 | 6.85e-108 | 751 | 1500 | 36 | 760 |
| SQI53822.1 | 4.99e-107 | 440 | 1507 | 49 | 1021 |
| AVV57259.1 | 1.79e-102 | 750 | 1499 | 220 | 949 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6F2P_A | 6.27e-94 | 746 | 1507 | 5 | 722 | Structureof Paenibacillus xanthan lyase to 2.6 A resolution [Paenibacillus] |
| 2E24_A | 7.17e-86 | 758 | 1482 | 11 | 700 | ChainA, Xanthan lyase [Bacillus sp. GL1] |
| 2E22_A | 3.23e-85 | 758 | 1482 | 11 | 700 | Crystalstructure of xanthan lyase in complex with mannose [Bacillus sp. GL1] |
| 1J0M_A | 7.96e-85 | 758 | 1482 | 11 | 700 | CrystalStructure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan [Bacillus sp. GL1],1J0N_A Crystal Structure of Bacillus sp. GL1 Xanthan Lyase that Acts on Side Chains of Xanthan [Bacillus sp. GL1] |
| 1X1H_A | 3.58e-84 | 758 | 1482 | 11 | 700 | ChainA, xanthan lyase [Bacillus sp. GL1],1X1I_A Chain A, xanthan lyase [Bacillus sp. GL1],1X1J_A Chain A, xanthan lyase [Bacillus sp. GL1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9AQS0 | 5.77e-83 | 758 | 1482 | 36 | 725 | Xanthan lyase OS=Bacillus sp. (strain GL1) OX=84635 GN=xly PE=1 SV=1 |
| Q59801 | 8.92e-71 | 751 | 1486 | 49 | 777 | Hyaluronate lyase OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=hysA PE=3 SV=1 |
| Q53591 | 4.03e-66 | 744 | 1404 | 245 | 897 | Hyaluronate lyase OS=Streptococcus agalactiae serotype III (strain NEM316) OX=211110 GN=hylB PE=1 SV=2 |
| Q54873 | 3.80e-57 | 745 | 1404 | 285 | 916 | Hyaluronate lyase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0314 PE=1 SV=2 |
| P0CZ01 | 4.44e-57 | 750 | 1491 | 38 | 757 | Hyaluronate lyase OS=Cutibacterium acnes (strain DSM 16379 / KPA171202) OX=267747 GN=PPA0380 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
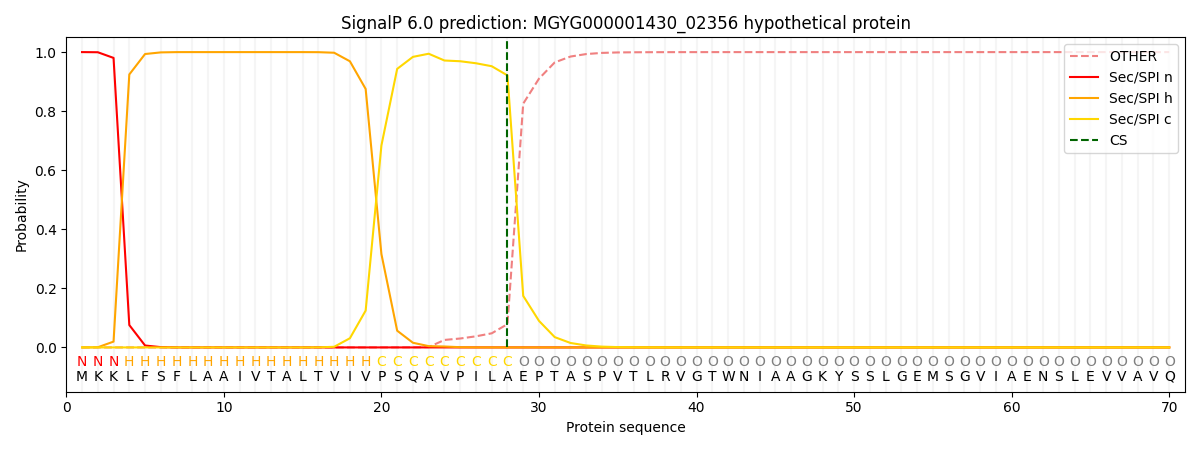
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000310 | 0.998900 | 0.000190 | 0.000203 | 0.000195 | 0.000172 |