You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001430_03395
You are here: Home > Sequence: MGYG000001430_03395
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Holdemania massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Holdemania; Holdemania massiliensis | |||||||||||
| CAZyme ID | MGYG000001430_03395 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 125637; End: 127565 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 229 | 453 | 4.3e-49 | 0.9147982062780269 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 3.74e-21 | 474 | 637 | 10 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 7.61e-16 | 473 | 641 | 507 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| pfam13231 | PMT_2 | 4.25e-15 | 256 | 400 | 1 | 136 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1928 | PMT1 | 2.44e-13 | 215 | 457 | 49 | 251 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG4346 | COG4346 | 3.21e-13 | 192 | 604 | 46 | 425 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANY70643.1 | 8.36e-123 | 34 | 639 | 432 | 1048 |
| QNK56024.1 | 1.08e-121 | 65 | 641 | 693 | 1271 |
| AIQ72362.1 | 2.91e-121 | 50 | 641 | 2 | 592 |
| BCN30459.1 | 3.12e-118 | 27 | 639 | 451 | 1056 |
| AIQ66842.1 | 3.76e-116 | 67 | 641 | 454 | 1031 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000047 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
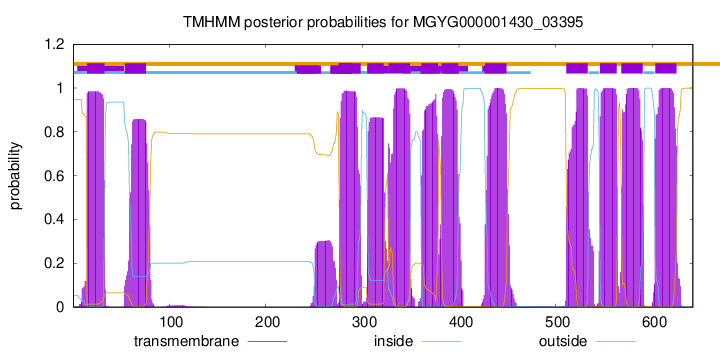
| start | end |
|---|---|
| 15 | 33 |
| 54 | 76 |
| 276 | 298 |
| 305 | 322 |
| 327 | 349 |
| 361 | 378 |
| 382 | 399 |
| 427 | 449 |
| 511 | 533 |
| 546 | 563 |
| 568 | 590 |
| 603 | 625 |
