You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001431_00065
You are here: Home > Sequence: MGYG000001431_00065
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
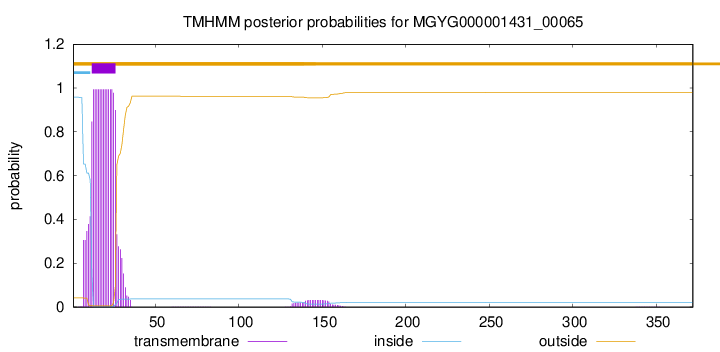
TMHMM annotations
Basic Information help
| Species | Helicobacter pylori_BU | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Helicobacteraceae; Helicobacter; Helicobacter pylori_BU | |||||||||||
| CAZyme ID | MGYG000001431_00065 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1610; End: 2728 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4692 | COG4692 | 2.91e-173 | 1 | 372 | 1 | 381 | Predicted neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam13088 | BNR_2 | 1.38e-73 | 74 | 344 | 1 | 275 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| cd15482 | Sialidase_non-viral | 7.61e-05 | 55 | 156 | 6 | 106 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEF24152.1 | 4.75e-267 | 1 | 372 | 1 | 372 |
| QEF43228.1 | 4.75e-267 | 1 | 372 | 1 | 372 |
| SMA52593.1 | 1.59e-265 | 1 | 372 | 1 | 372 |
| AVL49141.1 | 5.31e-264 | 1 | 372 | 1 | 372 |
| AKE81484.1 | 5.31e-264 | 1 | 372 | 1 | 372 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
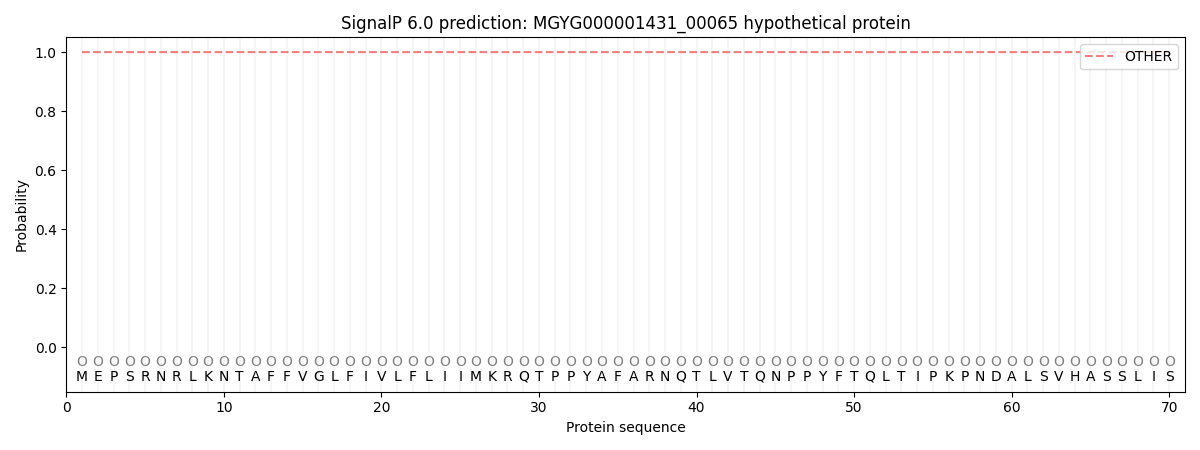
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000040 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |