You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001432_01112
Basic Information
help
| Species |
Helicobacter_C cinaedi
|
| Lineage |
Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Helicobacteraceae; Helicobacter_C; Helicobacter_C cinaedi
|
| CAZyme ID |
MGYG000001432_01112
|
| CAZy Family |
GH33 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 425 |
|
48114.79 |
7.1573 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001432 |
2240130 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 1087314;
End: 1088591
Strand: -
|
No EC number prediction in MGYG000001432_01112.
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam13088
|
BNR_2 |
5.24e-62 |
89 |
398 |
1 |
280 |
BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG4692
|
COG4692 |
5.02e-31 |
66 |
409 |
24 |
377 |
Predicted neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
This protein is predicted as SP
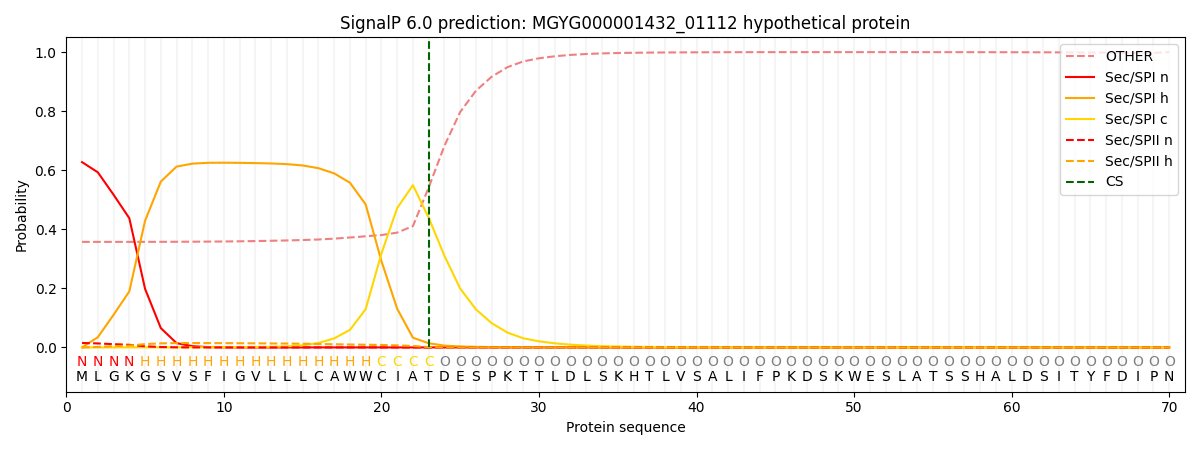
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.378941
|
0.602293
|
0.017166
|
0.000438
|
0.000369
|
0.000773
|
There is no transmembrane helices in MGYG000001432_01112.

