You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_01520
You are here: Home > Sequence: MGYG000001433_01520
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_01520 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1933169; End: 1937371 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 23 | 782 | 3.5e-284 | 0.9833795013850416 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 6.09e-75 | 29 | 278 | 1 | 233 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| PRK15319 | PRK15319 | 1.11e-05 | 838 | 1061 | 600 | 845 | fibronectin-binding autotransporter adhesin ShdA. |
| COG4354 | COG4354 | 6.94e-04 | 443 | 621 | 419 | 590 | Uncharacterized protein, contains GBA2_N and DUF608 domains [Function unknown]. |
| PRK15319 | PRK15319 | 0.002 | 882 | 1061 | 775 | 971 | fibronectin-binding autotransporter adhesin ShdA. |
| TIGR01414 | autotrans_barl | 0.003 | 1182 | 1259 | 1 | 87 | outer membrane autotransporter barrel domain. A number of Gram-negative bacterial proteins, mostly found in pathogens and associated with virulence, contain a conserved C-terminal domain that integrates into the outer membrane and enables the N-terminal region to be delivered across the membrane. This C-terminal autotransporter domain is about 400 amino acids in length and includes the aromatic amino acid-rich OMP signal, typically ending with a Phe or Trp residue, at the extreme C-terminus. [Protein fate, Protein and peptide secretion and trafficking, Cellular processes, Pathogenesis] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT73981.1 | 0.0 | 1 | 1400 | 1 | 1400 |
| QCP72031.1 | 0.0 | 22 | 843 | 18 | 841 |
| QCD38342.1 | 0.0 | 22 | 843 | 18 | 841 |
| AWK06253.1 | 0.0 | 21 | 845 | 21 | 844 |
| APA00322.1 | 0.0 | 21 | 845 | 17 | 839 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4UFC_A | 1.25e-259 | 17 | 843 | 12 | 802 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 7KMQ_A | 1.38e-184 | 23 | 783 | 37 | 760 | ChainA, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306],7KMQ_B Chain B, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306] |
| 2RDY_A | 7.85e-169 | 26 | 772 | 2 | 744 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 2EAB_A | 4.41e-95 | 20 | 802 | 12 | 867 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 2EAD_A | 1.53e-94 | 20 | 802 | 12 | 867 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 2.29e-155 | 16 | 793 | 41 | 830 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 2.57e-85 | 24 | 766 | 18 | 762 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q5AU81 | 5.86e-74 | 8 | 775 | 3 | 792 | Alpha-fucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=afcA PE=1 SV=1 |
| Q2USL3 | 1.14e-62 | 7 | 780 | 1 | 714 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
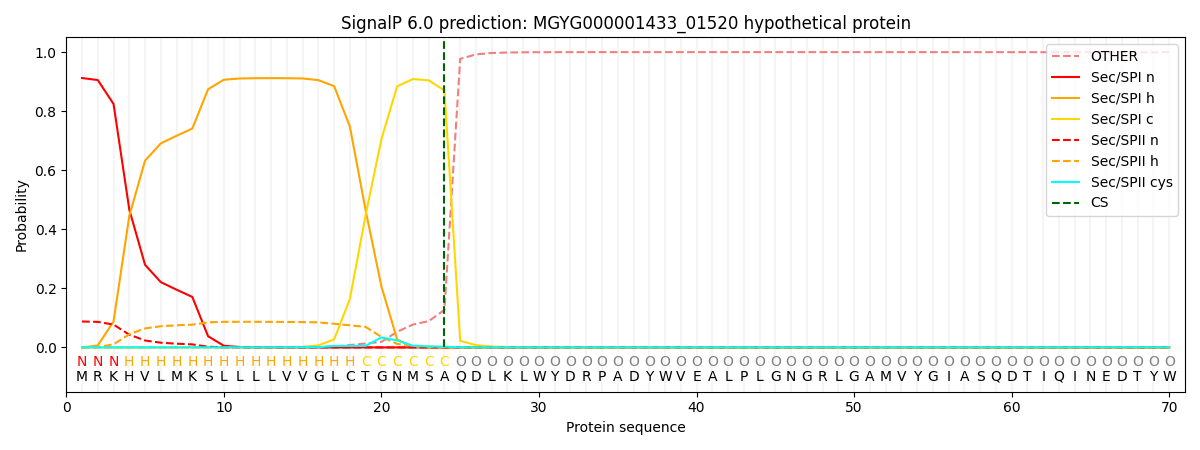
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001022 | 0.905939 | 0.092303 | 0.000251 | 0.000236 | 0.000228 |
