You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_01876
You are here: Home > Sequence: MGYG000001433_01876
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_01876 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2430571; End: 2432985 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 30 | 772 | 6.9e-226 | 0.9667590027700831 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14498 | Glyco_hyd_65N_2 | 2.89e-61 | 33 | 270 | 1 | 231 | Glycosyl hydrolase family 65, N-terminal domain. This domain represents a domain found to the N-terminus of the glycosyl hydrolase 65 family catalytic domain. |
| cd14771 | TrHb2_Mt-trHbO-like_O | 2.67e-05 | 451 | 504 | 22 | 88 | Truncated hemoglobins, group 2 (O); Mycobacterium tuberculosis hemoglobin O like. The M- and S families exhibit the canonical secondary structure of hemoglobins, a 3-over-3 alpha-helical sandwich structure (3/3 Mb-fold), built by eight alpha-helical segments. Truncated hemoglobins (TrHbs, 2/2Hb, or 2/2 globins) or the T family globins adopt a 2-on-2 alpha-helical sandwich structure, resulting from extensive and complex modifications of the canonical 3-on-3 alpha-helical sandwich that are distributed throughout the whole protein molecule. TrHbs are classified into three main groups based on their structural properties and named after Mycobacterium sp. genes glbN, glbO, and glbP: TrHb1s (N), TrHb2s (O) and TrHb3s (P). This group includes a Mycobacterium tuberculosis TrHb2, Mt-trHbO, encoded by the Mycobacterium tuberculosis glbO gene, which is expressed throughout the Mycobacterium growth phase. It also includes a TrHb2 from the thermophilic Thermobifida fusca ( Tf-trHb) which has a high thermostability and at the optimal growth temperature for Thermobifida fusca (between 55 and 60 degrees C ), it is capable of efficient O2 binding and release. Tf-trHb shares a relatively slow rate of oxygen binding with Mt-trHbO. |
| COG1554 | ATH1 | 2.32e-04 | 424 | 509 | 384 | 479 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74335.1 | 0.0 | 1 | 804 | 1 | 804 |
| BCI62991.1 | 0.0 | 2 | 794 | 7 | 795 |
| QEK99247.1 | 6.11e-170 | 22 | 779 | 19 | 770 |
| QEL85753.1 | 1.36e-166 | 32 | 776 | 48 | 759 |
| AZQ64685.1 | 5.20e-166 | 16 | 769 | 290 | 1028 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RDY_A | 8.87e-141 | 35 | 770 | 7 | 743 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 4UFC_A | 6.11e-126 | 33 | 769 | 24 | 731 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 7KMQ_A | 1.46e-117 | 33 | 769 | 43 | 747 | ChainA, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306],7KMQ_B Chain B, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306] |
| 2EAD_A | 2.33e-93 | 28 | 782 | 15 | 851 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum],2EAD_B Chain B, Alpha-fucosidase [Bifidobacterium bifidum] |
| 2EAB_A | 1.20e-92 | 28 | 782 | 15 | 851 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 6.60e-127 | 25 | 766 | 46 | 799 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
| A2R797 | 3.77e-99 | 31 | 759 | 21 | 756 | Probable alpha-fucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=afcA PE=3 SV=1 |
| Q5AU81 | 5.75e-70 | 18 | 759 | 12 | 775 | Alpha-fucosidase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=afcA PE=1 SV=1 |
| Q2USL3 | 4.36e-55 | 27 | 787 | 12 | 722 | Probable alpha-fucosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=afcA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
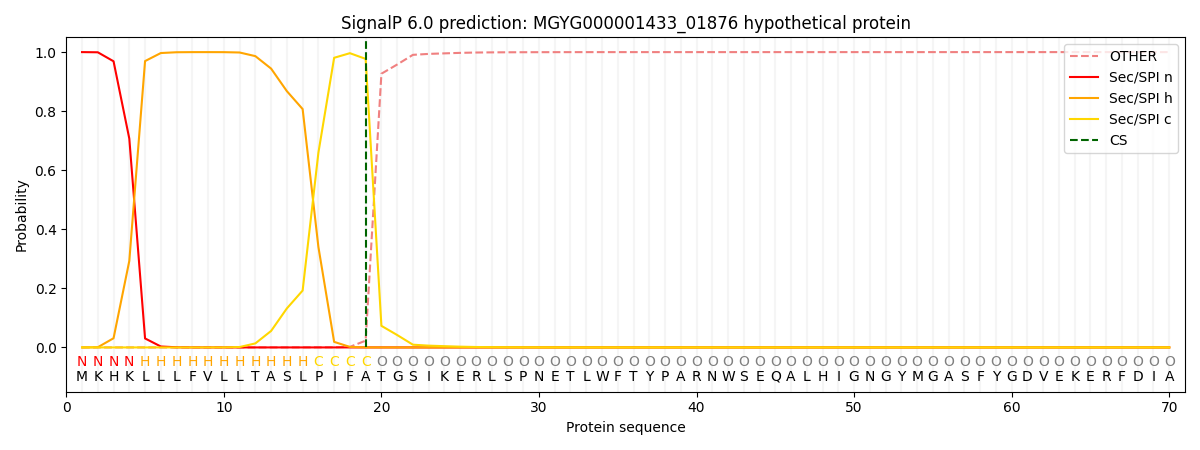
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000464 | 0.998756 | 0.000248 | 0.000178 | 0.000169 | 0.000158 |
