You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_01978
You are here: Home > Sequence: MGYG000001433_01978
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_01978 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2601267; End: 2603237 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 297 | 576 | 3.7e-17 | 0.6496519721577726 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.00e-06 | 244 | 472 | 29 | 239 | Cellulase (glycosyl hydrolase family 5). |
| pfam00754 | F5_F8_type_C | 9.64e-06 | 52 | 191 | 6 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam01229 | Glyco_hydro_39 | 3.39e-04 | 231 | 474 | 23 | 276 | Glycosyl hydrolases family 39. |
| COG3867 | GanB | 0.001 | 244 | 429 | 68 | 245 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74462.1 | 0.0 | 1 | 656 | 1 | 656 |
| ANE45240.1 | 4.40e-45 | 213 | 653 | 33 | 468 |
| QJD87872.1 | 1.73e-44 | 215 | 597 | 738 | 1113 |
| QGQ99779.1 | 1.99e-16 | 60 | 200 | 590 | 717 |
| QUI21585.1 | 1.47e-14 | 39 | 203 | 956 | 1136 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
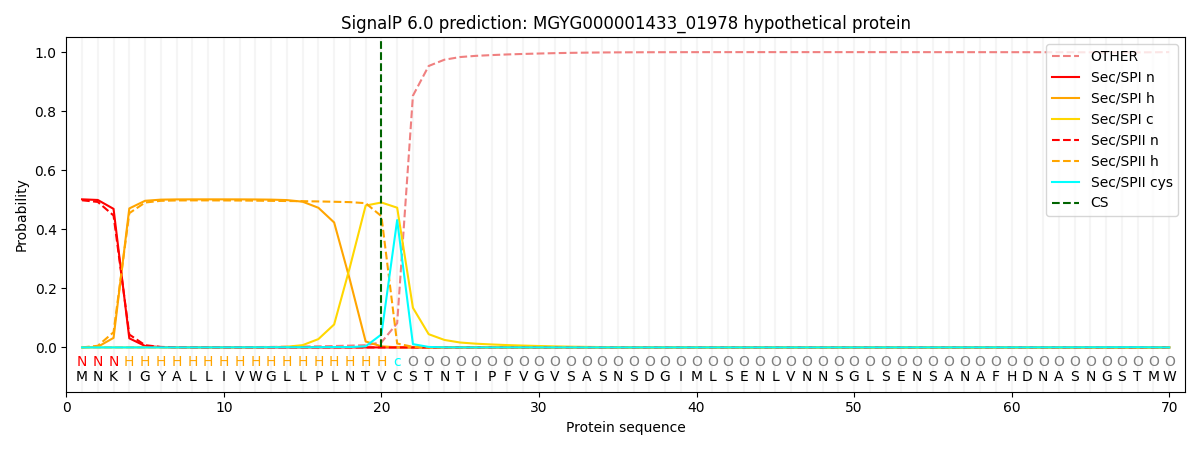
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001393 | 0.495173 | 0.502944 | 0.000158 | 0.000161 | 0.000161 |
