You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_03014
You are here: Home > Sequence: MGYG000001433_03014
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_03014 | |||||||||||
| CAZy Family | GT26 | |||||||||||
| CAZyme Description | Undecaprenyl-phosphate alpha-N-acetylglucosaminyl 1-phosphate transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 801217; End: 802986 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT26 | 385 | 557 | 1.1e-50 | 0.9883040935672515 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06853 | GT_WecA_like | 7.62e-77 | 5 | 271 | 1 | 249 | This subfamily contains Escherichia coli WecA, Bacillus subtilis TagO and related proteins. WecA is an UDP-N-acetylglucosamine (GlcNAc):undecaprenyl-phosphate (Und-P) GlcNAc-1-phosphate transferase that catalyzes the formation of a phosphodiester bond between a membrane-associated undecaprenyl-phosphate molecule and N-acetylglucosamine 1-phosphate, which is usually donated by a soluble UDP-N-acetylglucosamine precursor. WecA participates in the biosynthesis of O antigen LPS in many enteric bacteria and is also involved in the biosynthesis of enterobacterial common antigen. A conserved short sequence motif and a conserved arginine at a cytosolic loop of this integral membrane protein were shown to be critical in recognition of substrate UDP-N-acetylglucosamine. |
| pfam03808 | Glyco_tran_WecB | 7.15e-66 | 388 | 557 | 1 | 168 | Glycosyl transferase WecB/TagA/CpsF family. |
| cd06533 | Glyco_transf_WecG_TagA | 2.30e-60 | 385 | 557 | 1 | 170 | The glycosyltransferase WecG/TagA superfamily contains Escherichia coli WecG, Bacillus subtilis TagA and related proteins. E. coli WecG is believed to be a UDP-N-acetyl-D-mannosaminuronic acid transferase, and is involved in enterobacterial common antigen (eca) synthesis. B. subtilis TagA plays a key role in the Wall Teichoic Acid (WTA) biosynthetic pathway, catalyzing the transfer of N-acetylmannosamine to the C4 hydroxyl of a membrane-anchored N-acetylglucosaminyl diphospholipid to make ManNAc-beta-(1,4)-GlcNAc-pp-undecaprenyl. This is the first committed step in this pathway. Also included in this group is Xanthomonas campestris pv. campestris GumM, a glycosyltransferase participating in the biosynthesis of the exopolysaccharide xanthan. |
| COG1922 | WecG | 3.74e-55 | 359 | 566 | 37 | 241 | UDP-N-acetyl-D-mannosaminuronic acid transferase, WecB/TagA/CpsF family [Cell wall/membrane/envelope biogenesis]. |
| COG0472 | Rfe | 8.05e-45 | 3 | 296 | 38 | 314 | UDP-N-acetylmuramyl pentapeptide phosphotransferase/UDP-N-acetylglucosamine-1-phosphate transferase [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT75934.1 | 0.0 | 1 | 589 | 1 | 589 |
| QCT93965.1 | 3.03e-81 | 359 | 574 | 22 | 237 |
| AMQ57519.1 | 4.32e-76 | 357 | 574 | 16 | 234 |
| AEI14038.1 | 5.09e-76 | 341 | 573 | 5 | 237 |
| QJD98253.1 | 5.65e-74 | 360 | 575 | 17 | 232 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7MPK_A | 1.31e-23 | 363 | 570 | 34 | 236 | ChainA, N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [Thermoanaerobacter italicus Ab9],7MPK_B Chain B, N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [Thermoanaerobacter italicus Ab9],7MPK_C Chain C, N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [Thermoanaerobacter italicus Ab9],7N41_A Chain A, N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [Thermoanaerobacter italicus Ab9],7N41_B Chain B, N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [Thermoanaerobacter italicus Ab9],7N41_C Chain C, N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase [Thermoanaerobacter italicus Ab9] |
| 5WB4_A | 1.52e-17 | 363 | 527 | 34 | 195 | Crystalstructure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_B Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_C Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_D Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_E Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_F Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_G Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9],5WB4_H Crystal structure of the TarA wall teichoic acid glycosyltransferase [Thermoanaerobacter italicus Ab9] |
| 5WFG_A | 2.80e-17 | 363 | 527 | 34 | 195 | Crystalstructure of the TarA wall teichoic acid glycosyltransferase bound to UDP [Thermoanaerobacter italicus Ab9],5WFG_B Crystal structure of the TarA wall teichoic acid glycosyltransferase bound to UDP [Thermoanaerobacter italicus Ab9],5WFG_C Crystal structure of the TarA wall teichoic acid glycosyltransferase bound to UDP [Thermoanaerobacter italicus Ab9],5WFG_D Crystal structure of the TarA wall teichoic acid glycosyltransferase bound to UDP [Thermoanaerobacter italicus Ab9],5WFG_E Crystal structure of the TarA wall teichoic acid glycosyltransferase bound to UDP [Thermoanaerobacter italicus Ab9],5WFG_F Crystal structure of the TarA wall teichoic acid glycosyltransferase bound to UDP [Thermoanaerobacter italicus Ab9] |
| 4J72_A | 2.24e-10 | 9 | 296 | 76 | 363 | CrystalStructure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase [Aquifex aeolicus],4J72_B Crystal Structure of polyprenyl-phosphate N-acetyl hexosamine 1-phosphate transferase [Aquifex aeolicus],5CKR_A Crystal Structure of MraY in complex with Muraymycin D2 [Aquifex aeolicus VF5],6OYH_A Crystal structure of MraY bound to carbacaprazamycin [Aquifex aeolicus VF5],6OYH_B Crystal structure of MraY bound to carbacaprazamycin [Aquifex aeolicus VF5],6OYH_C Crystal structure of MraY bound to carbacaprazamycin [Aquifex aeolicus VF5],6OYH_D Crystal structure of MraY bound to carbacaprazamycin [Aquifex aeolicus VF5],6OYZ_A Crystal structure of MraY bound to capuramycin [Aquifex aeolicus VF5],6OYZ_B Crystal structure of MraY bound to capuramycin [Aquifex aeolicus VF5],6OYZ_C Crystal structure of MraY bound to capuramycin [Aquifex aeolicus VF5],6OYZ_D Crystal structure of MraY bound to capuramycin [Aquifex aeolicus VF5],6OZ6_A Crystal structure of MraY bound to 3'-hydroxymureidomycin A [Aquifex aeolicus VF5],6OZ6_B Crystal structure of MraY bound to 3'-hydroxymureidomycin A [Aquifex aeolicus VF5],6OZ6_C Crystal structure of MraY bound to 3'-hydroxymureidomycin A [Aquifex aeolicus VF5],6OZ6_D Crystal structure of MraY bound to 3'-hydroxymureidomycin A [Aquifex aeolicus VF5] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34753 | 9.26e-33 | 2 | 280 | 40 | 297 | Probable undecaprenyl-phosphate N-acetylglucosaminyl 1-phosphate transferase OS=Bacillus subtilis (strain 168) OX=224308 GN=tagO PE=1 SV=1 |
| P27620 | 2.34e-29 | 363 | 582 | 35 | 243 | N-acetylglucosaminyldiphosphoundecaprenol N-acetyl-beta-D-mannosaminyltransferase OS=Bacillus subtilis (strain 168) OX=224308 GN=tagA PE=1 SV=1 |
| P9WMW4 | 3.98e-28 | 2 | 280 | 64 | 350 | Decaprenyl-phosphate N-acetylglucosaminephosphotransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=wecA PE=3 SV=1 |
| P9WMW5 | 3.98e-28 | 2 | 280 | 64 | 350 | Decaprenyl-phosphate N-acetylglucosaminephosphotransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=wecA PE=1 SV=1 |
| P45830 | 6.70e-28 | 2 | 280 | 64 | 350 | Decaprenyl-phosphate N-acetylglucosaminephosphotransferase OS=Mycobacterium leprae (strain TN) OX=272631 GN=wecA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
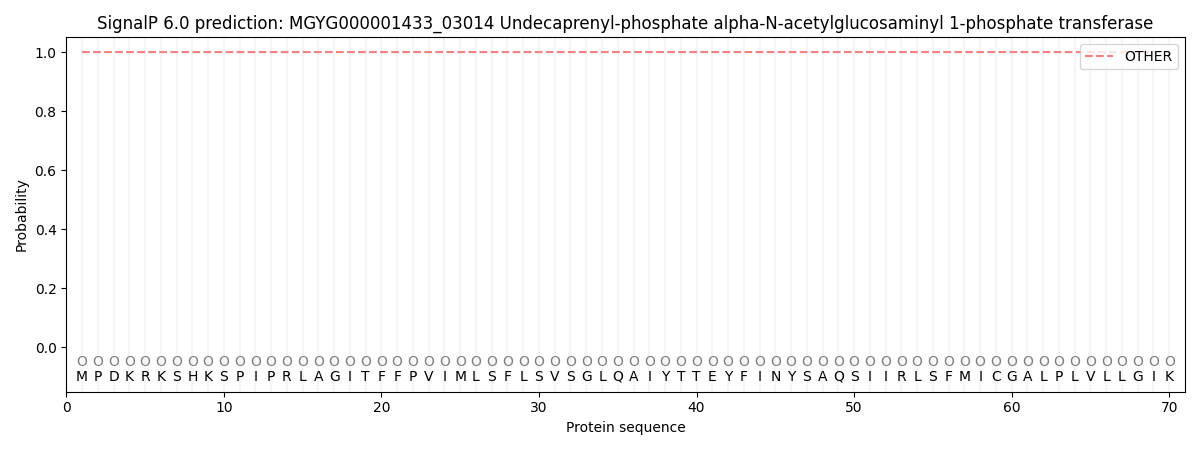
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000031 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help

| start | end |
|---|---|
| 15 | 37 |
| 50 | 67 |
| 82 | 99 |
| 106 | 128 |
| 138 | 160 |
| 165 | 184 |
| 189 | 211 |
| 216 | 238 |
| 276 | 295 |
| 300 | 318 |
