You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_03658
You are here: Home > Sequence: MGYG000001433_03658
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_03658 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1527574; End: 1529193 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 39 | 208 | 6.3e-21 | 0.46089385474860334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR04183 | Por_Secre_tail | 2.29e-04 | 469 | 538 | 1 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| pfam07221 | GlcNAc_2-epim | 0.003 | 124 | 273 | 149 | 337 | N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase). This family contains a number of eukaryotic and bacterial N-acylglucosamine 2-epimerase (GlcNAc 2-epimerase) enzymes (EC:5.3.1.8) approximately 500 residues long. This converts N-acyl-D-glucosamine to N-acyl-D-mannosamine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADO68190.1 | 3.22e-94 | 25 | 456 | 225 | 643 |
| ATB43874.1 | 7.94e-91 | 17 | 456 | 203 | 629 |
| ATB28105.1 | 1.13e-87 | 17 | 456 | 189 | 611 |
| ARN56013.1 | 4.14e-67 | 17 | 451 | 22 | 459 |
| AQQ08789.1 | 9.87e-62 | 17 | 451 | 22 | 459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4V1S_A | 6.36e-24 | 16 | 345 | 37 | 388 | Structureof the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4V1S_B Structure of the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4V1R_A | 3.10e-22 | 16 | 345 | 37 | 388 | Structureof a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4V1R_B Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4MU9_A | 1.41e-14 | 98 | 319 | 103 | 339 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 6U4Z_A | 2.01e-13 | 89 | 207 | 207 | 325 | CrystalStructure of a family 76 glycoside hydrolase from a bovine Bacteroides thetaiotaomicron strain [Bacteroides thetaiotaomicron] |
| 4C1S_A | 3.68e-13 | 89 | 206 | 83 | 200 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
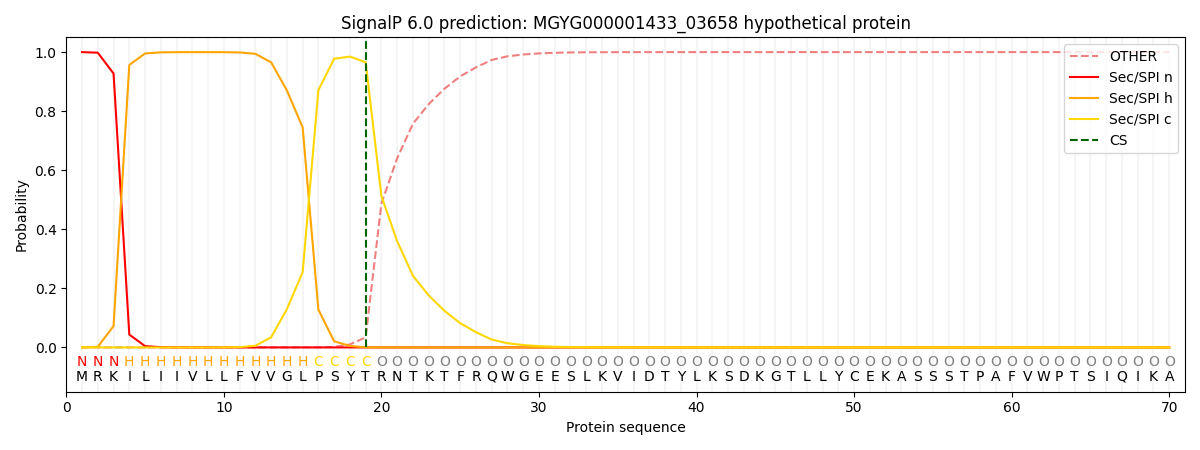
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001250 | 0.997657 | 0.000452 | 0.000217 | 0.000205 | 0.000201 |
