You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001433_04065
You are here: Home > Sequence: MGYG000001433_04065
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides salyersiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides salyersiae | |||||||||||
| CAZyme ID | MGYG000001433_04065 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 133246; End: 139878 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 2010 | 2117 | 5e-20 | 0.8548387096774194 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.27e-14 | 2012 | 2116 | 13 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam13472 | Lipase_GDSL_2 | 9.06e-12 | 751 | 922 | 2 | 175 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam00754 | F5_F8_type_C | 1.45e-08 | 1531 | 1630 | 16 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| TIGR04183 | Por_Secre_tail | 8.10e-07 | 2142 | 2210 | 1 | 72 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| cd01834 | SGNH_hydrolase_like_2 | 1.22e-06 | 747 | 931 | 3 | 191 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT74836.1 | 0.0 | 1 | 2210 | 1 | 2210 |
| QHS63397.1 | 2.30e-130 | 246 | 1130 | 26 | 824 |
| AUB37190.1 | 1.51e-20 | 744 | 934 | 131 | 319 |
| BBD67944.1 | 7.94e-20 | 744 | 934 | 131 | 319 |
| ACC82951.1 | 3.09e-19 | 742 | 934 | 127 | 317 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
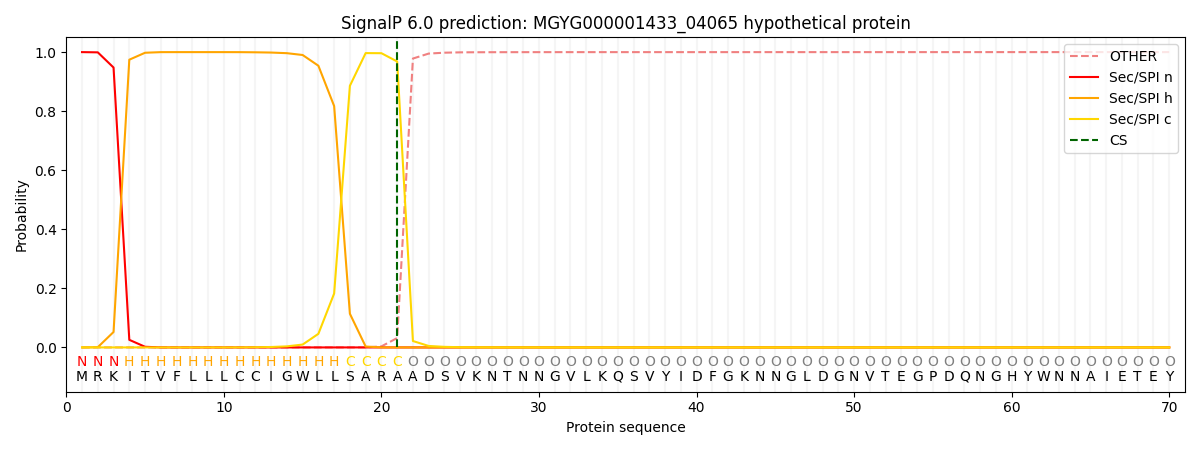
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000305 | 0.998928 | 0.000243 | 0.000162 | 0.000159 | 0.000148 |
