You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_00092
You are here: Home > Sequence: MGYG000001436_00092
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_F sp000411255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255 | |||||||||||
| CAZyme ID | MGYG000001436_00092 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 132193; End: 133734 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 32 | 406 | 4.6e-149 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05048 | NosD | 2.51e-06 | 200 | 421 | 2 | 182 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| pfam13229 | Beta_helix | 4.04e-05 | 224 | 395 | 7 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| cd14251 | PL-6 | 6.02e-05 | 56 | 419 | 7 | 367 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam08480 | Disaggr_assoc | 0.005 | 226 | 440 | 2 | 187 | Disaggregatase related. This domain is found in disaggregatases and several hypothetical proteins of the archaeal genus Methanosarcina. Disaggregatases cause aggregates to separate into single cells and contain parallel beta-helix repeats. Also see pfam06848. |
| pfam13229 | Beta_helix | 0.006 | 289 | 421 | 20 | 137 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV19466.1 | 0.0 | 1 | 513 | 1 | 514 |
| QLG40737.1 | 6.01e-296 | 1 | 513 | 2 | 515 |
| QOS78982.1 | 8.13e-294 | 1 | 513 | 2 | 515 |
| QQZ61545.1 | 1.43e-289 | 1 | 513 | 2 | 514 |
| CQR57779.1 | 1.40e-285 | 1 | 513 | 2 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VST_A | 1.03e-06 | 34 | 108 | 2 | 76 | Thecomplex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_B The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_C The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_D The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_A The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_B The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_C The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_D The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_A The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_B The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_C The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_D The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94576 | 5.76e-102 | 31 | 480 | 32 | 463 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
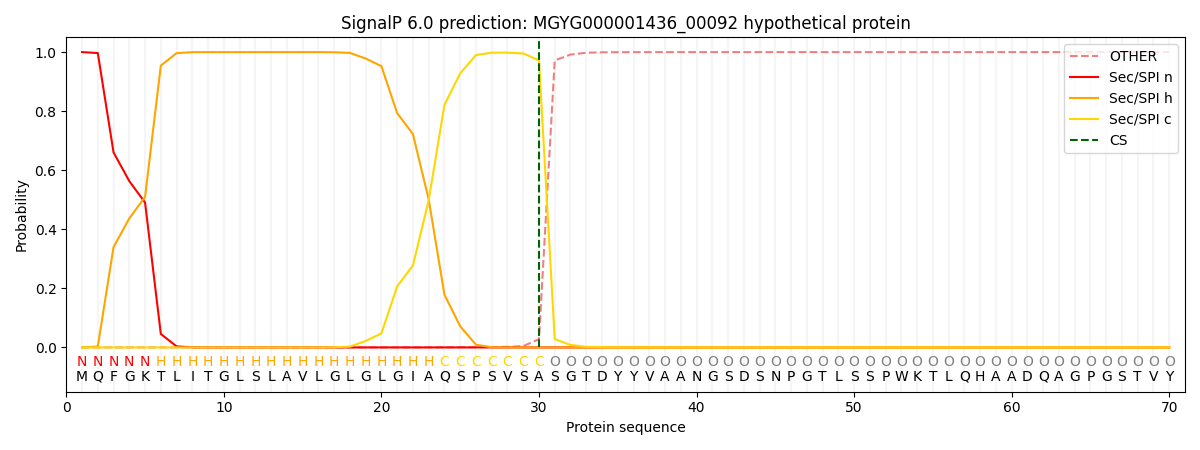
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000242 | 0.999094 | 0.000179 | 0.000177 | 0.000156 | 0.000140 |
