You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_00382
You are here: Home > Sequence: MGYG000001436_00382
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
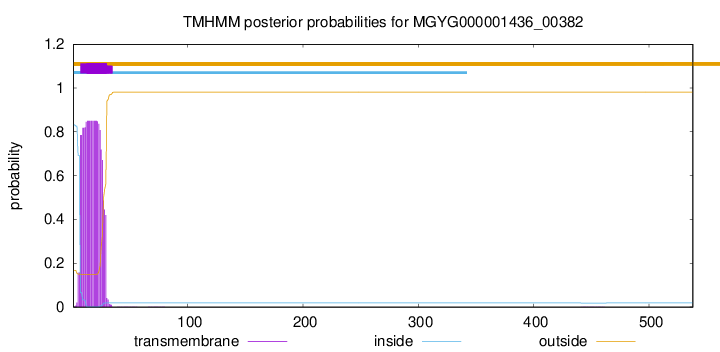
TMHMM annotations
Basic Information help
| Species | Paenibacillus_F sp000411255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255 | |||||||||||
| CAZyme ID | MGYG000001436_00382 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 489567; End: 491183 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 186 | 300 | 5.7e-20 | 0.8467741935483871 |
| CBM32 | 45 | 168 | 8e-16 | 0.9354838709677419 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04450 | BSP | 3.00e-37 | 324 | 532 | 6 | 205 | Peptidase of plants and bacteria. These basic secretory proteins (BSPs) are believed to be part of the plants defense mechanism against pathogens. |
| pfam00754 | F5_F8_type_C | 7.90e-11 | 184 | 263 | 1 | 82 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 5.07e-09 | 45 | 168 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.03e-04 | 185 | 300 | 14 | 134 | Substituted updates: Jan 31, 2002 |
| COG0501 | HtpX | 0.003 | 338 | 433 | 87 | 172 | Zn-dependent protease with chaperone function [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV19752.1 | 0.0 | 1 | 538 | 1 | 537 |
| AJS58459.1 | 1.89e-241 | 1 | 538 | 1 | 530 |
| SYX86287.1 | 7.97e-219 | 1 | 538 | 1 | 530 |
| SYX85130.1 | 1.97e-207 | 1 | 538 | 1 | 530 |
| CQR57408.1 | 1.14e-179 | 1 | 536 | 2 | 528 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FGY9 | 4.98e-13 | 38 | 171 | 575 | 717 | Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase OS=Arabidopsis thaliana OX=3702 GN=PNG1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
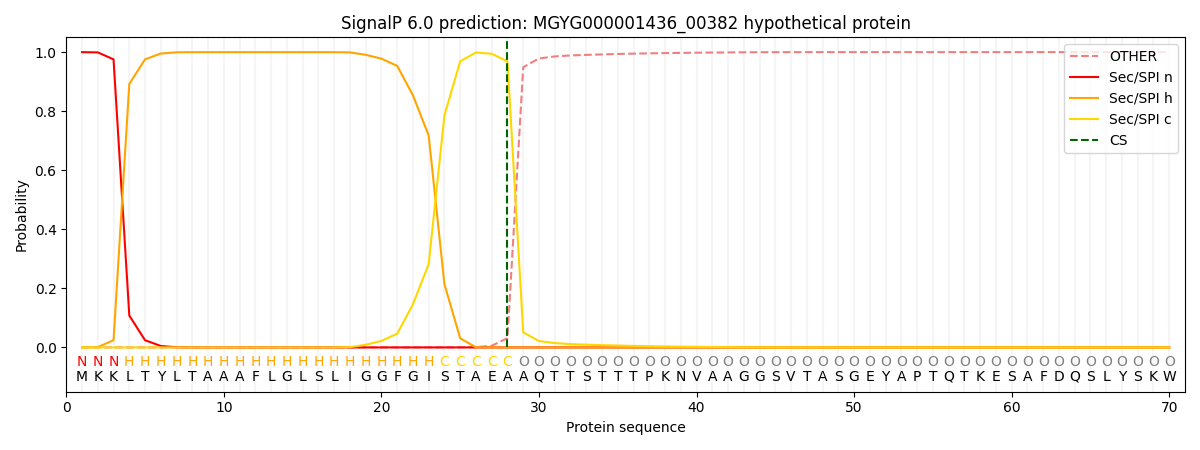
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000346 | 0.998749 | 0.000361 | 0.000204 | 0.000160 | 0.000141 |