You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_01150
You are here: Home > Sequence: MGYG000001436_01150
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_F sp000411255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255 | |||||||||||
| CAZyme ID | MGYG000001436_01150 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1295690; End: 1297906 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 603 | 736 | 9.8e-51 | 0.9925373134328358 |
| CBM32 | 466 | 592 | 1.1e-21 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 1.55e-62 | 602 | 737 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.35e-53 | 602 | 736 | 4 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam13402 | Peptidase_M60 | 1.52e-48 | 154 | 416 | 1 | 268 | Peptidase M60, enhancin and enhancin-like. This family of peptidases contains a zinc metallopeptidase motif (HEXXHX(8,28)E) and possesses mucinase activity. It includes the viral enhancins as well as enhancin-like peptidases from bacterial species. Enhancins are a class of metalloproteases found in some baculoviruses that enhance viral infection by degrading the peritrophic membrane (PM) of the insect midgut. Bacterial enhancins are found to be cytotoxic when compared to viral enhancin, however, suggesting that the bacterial enhancins do not enhance infection in the same way as viral enhancin. Bacterial enhancins may have evolved a distinct biochemical function. These bacterial domains are peptidases targetting host glycoproteins and thus probably play an important role in successful colonisation of both vertebrate mucosal surfaces and the invertebrate digestive tract by both mutualistic and pathogenic microbes. This family has been augmented by a merge with the sequences in the Enhancin Pfam family. |
| pfam00754 | F5_F8_type_C | 2.24e-19 | 465 | 592 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam17291 | M60-like_N | 1.32e-14 | 64 | 151 | 5 | 106 | N-terminal domain of M60-like peptidases. This accessory domain has a jelly roll topology. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV20527.1 | 0.0 | 1 | 738 | 1 | 738 |
| SMF85702.1 | 0.0 | 1 | 738 | 1 | 745 |
| AIQ68876.1 | 0.0 | 1 | 738 | 1 | 738 |
| QOS79680.1 | 0.0 | 1 | 738 | 1 | 744 |
| AJS58516.1 | 0.0 | 1 | 738 | 1 | 737 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EV7_A | 6.21e-91 | 35 | 449 | 3 | 412 | Thecrystal structure of a functionally unknown conserved protein mutant from Bacillus anthracis str. Ames [Bacillus anthracis str. Ames] |
| 4FCA_A | 3.52e-89 | 35 | 449 | 3 | 412 | Thecrystal structure of a functionally unknown conserved protein from Bacillus anthracis str. Ames. [Bacillus anthracis str. Ames] |
| 7JS4_A | 1.04e-39 | 51 | 736 | 25 | 752 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 2VMG_A | 5.63e-34 | 585 | 736 | 1 | 156 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2VMH_A | 8.80e-34 | 602 | 736 | 8 | 150 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0TR53 | 4.39e-23 | 464 | 594 | 629 | 765 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
| Q8XL08 | 4.39e-23 | 464 | 594 | 629 | 765 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagJ PE=1 SV=1 |
| Q6QR59 | 7.63e-15 | 74 | 430 | 530 | 894 | TRPM8 channel-associated factor 3 OS=Mus musculus OX=10090 GN=Tcaf3 PE=1 SV=1 |
| Q6P6V7 | 9.05e-14 | 82 | 430 | 538 | 894 | TRPM8 channel-associated factor 3 OS=Rattus norvegicus OX=10116 GN=Tcaf3 PE=2 SV=1 |
| A9WNA0 | 1.10e-13 | 587 | 737 | 1145 | 1304 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
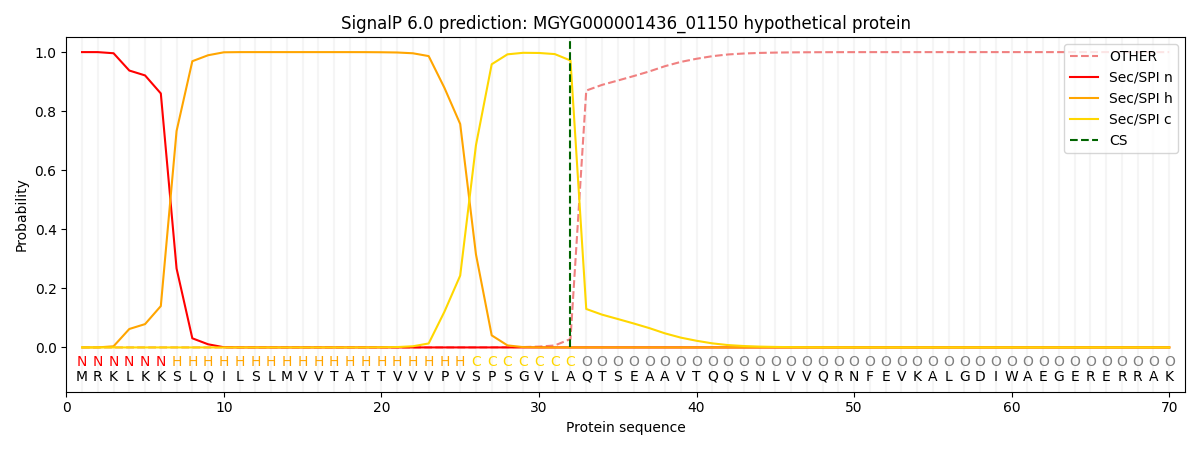
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000295 | 0.999010 | 0.000180 | 0.000177 | 0.000157 | 0.000142 |
