You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001436_02475
You are here: Home > Sequence: MGYG000001436_02475
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_F sp000411255 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_F; Paenibacillus_F sp000411255 | |||||||||||
| CAZyme ID | MGYG000001436_02475 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2780057; End: 2785111 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 479 | 809 | 2.9e-80 | 0.9459459459459459 |
| GH18 | 1342 | 1670 | 1.7e-78 | 0.9459459459459459 |
| GH18 | 911 | 1241 | 1.3e-77 | 0.9425675675675675 |
| CBM54 | 231 | 336 | 4.5e-29 | 0.8859649122807017 |
| SLH | 110 | 150 | 3.6e-16 | 0.9523809523809523 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06548 | GH18_chitinase | 3.65e-120 | 482 | 807 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| cd06548 | GH18_chitinase | 2.87e-116 | 913 | 1239 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| cd06548 | GH18_chitinase | 1.05e-111 | 1345 | 1668 | 1 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| smart00636 | Glyco_18 | 3.21e-100 | 481 | 807 | 1 | 334 | Glyco_18 domain. |
| smart00636 | Glyco_18 | 4.39e-99 | 912 | 1239 | 1 | 334 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAV16301.1 | 0.0 | 15 | 1684 | 1 | 1669 |
| BAM67143.1 | 0.0 | 25 | 1253 | 10 | 1415 |
| BBH23221.1 | 4.23e-111 | 50 | 443 | 29 | 430 |
| ALS25414.1 | 7.36e-111 | 19 | 446 | 3 | 426 |
| QGQ95647.1 | 1.64e-109 | 27 | 446 | 11 | 430 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GZT_B | 4.73e-232 | 297 | 1253 | 1 | 1133 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
| 5GZU_A | 4.93e-211 | 477 | 1253 | 23 | 882 | CrystalStructure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7],5GZU_B Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7],5GZV_A Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7],5GZV_B Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery [Paenibacillus sp. FPU-7] |
| 6INX_A | 1.22e-91 | 502 | 821 | 26 | 343 | Structuralinsights into a novel glycoside hydrolase family 18 N-acetylglucosaminidase from Paenibacillus barengoltzii [Paenibacillus barengoltzii] |
| 1ITX_A | 3.95e-71 | 481 | 816 | 13 | 415 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
| 6KST_A | 1.61e-70 | 912 | 1252 | 8 | 370 | Crystalstructure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KST_B Crystal structure of the catalytic domain of chitinase ChiL from Chitiniphilus shinanonensis (CsChiL) [Chitiniphilus shinanonensis],6KXL_A Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis],6KXL_B Crystal structure of the catalytic domain of Chitiniphilus shinanonensis chitinase ChiL (CsChiL) complexed with N,N'-diacetylchitobiose [Chitiniphilus shinanonensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P20533 | 1.72e-70 | 481 | 907 | 45 | 529 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| Q9W5U2 | 3.51e-58 | 908 | 1668 | 962 | 1753 | Probable chitinase 10 OS=Drosophila melanogaster OX=7227 GN=Cht10 PE=2 SV=2 |
| Q13231 | 2.12e-47 | 1344 | 1668 | 23 | 363 | Chitotriosidase-1 OS=Homo sapiens OX=9606 GN=CHIT1 PE=1 SV=1 |
| E9ERT9 | 1.26e-46 | 476 | 830 | 30 | 405 | Endochitinase 1 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chit1 PE=3 SV=1 |
| Q6RY07 | 6.77e-46 | 1367 | 1675 | 52 | 373 | Acidic mammalian chitinase OS=Rattus norvegicus OX=10116 GN=Chia PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
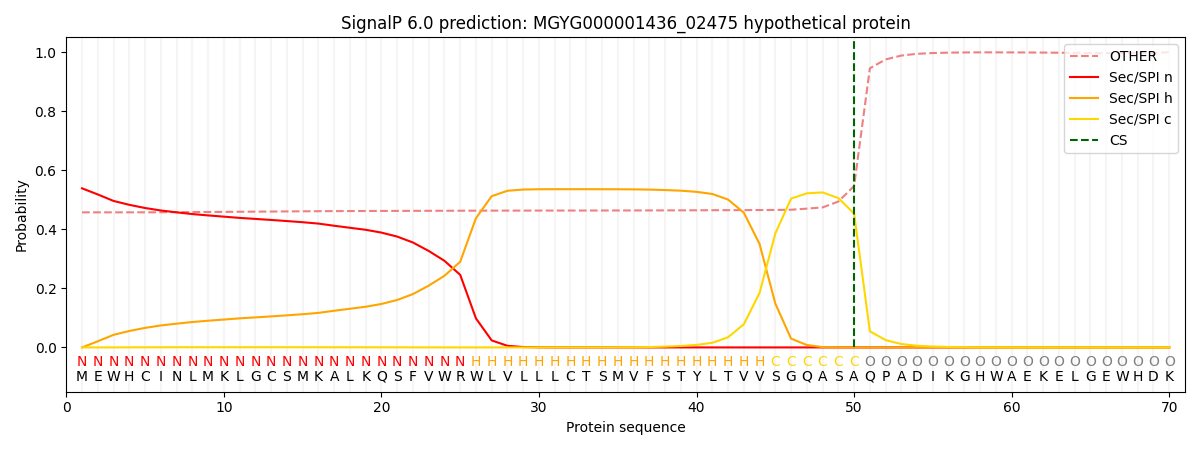
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.474913 | 0.516856 | 0.005068 | 0.001048 | 0.000593 | 0.001506 |

